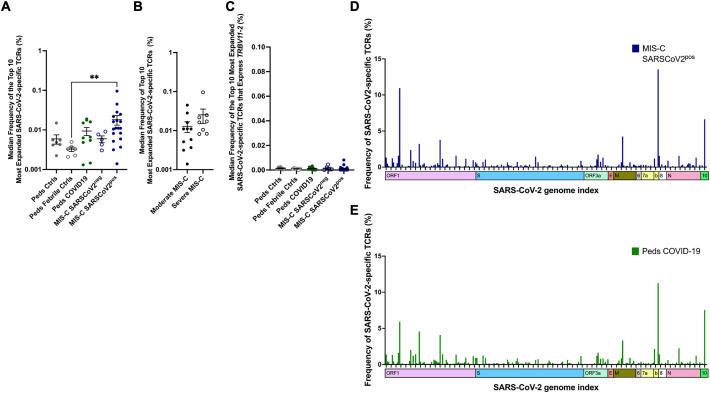
Fig. 6.
SARS-CoV-2-Specific T Cell Receptors in MIS-C. SARS-CoV-2-specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs for each patient in each study group. Statistical analysis Kruskal-Wallis test with Dunn's multiple comparison test. B) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs in patients with moderate and severe MIS-C. Statistical analysis: Mann-Whitney test C) The median frequency of the top 10 most abundant SARS-CoV-2-specific TCRs that express TRBV11–2 in each patient and each study group. Statistical analysis Kruskal-Wallis test with Dunn's multiple comparison test. D-E) Viral antigens recognized by SARS-CoV-2-specific TCRs in D) MIS-C SARSCOV2pos and E) pediatric COVID-19. Statistical analysis by permutation test with no difference between MIS-C SARSCOV2pos and peds COVID-19.
P value, ** ≤ 0.01.
Summary data on bar graphs is mean ± standard error.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; TCR, T cell receptor.