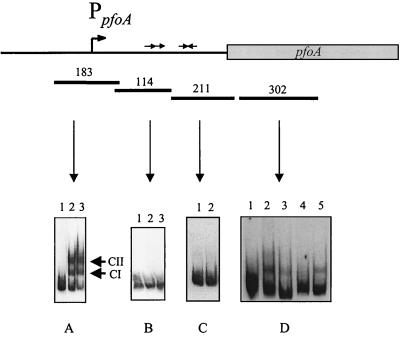
FIG. 3.
Gel shift analysis of the pfoA gene region. The pfoA-derived DNA fragments used in the gel mobility shift assays (bars) and their respective sizes (in base pairs) are shown. The direct repeats in the 114-bp fragment, the promoter in the 183-bp fragment, and the inverted repeats in the 211-bp fragment, are shown as directly repeated arrows, a bent arrow (PpfoA), and inverted arrows, respectively. (A and B) Lane 1, no-VirR control; lanes 2 and 3, DIG-labelled DNA incubated with 1 and 2 μg of VirR, respectively. The VirR-DNA complexes CI and CII are indicated. (C) Lane 1, no-VirR control; lane 2, DIG-labelled fragment incubated with 1 μg of VirR. (D) Lane 1, no-VirR control; lanes 2 to 5, target DNA incubated with 1 μg of VirR. Reaction mixtures in lanes 3 to 5 also contained 15 pmol of the following unlabelled fragments: lane 3, 302-bp fragment (specific competitor); lane 4, 183-bp of unlabelled upstream pfoA fragment; lane 5, 408-bp upstream pfoR fragment (nonspecific competitor).