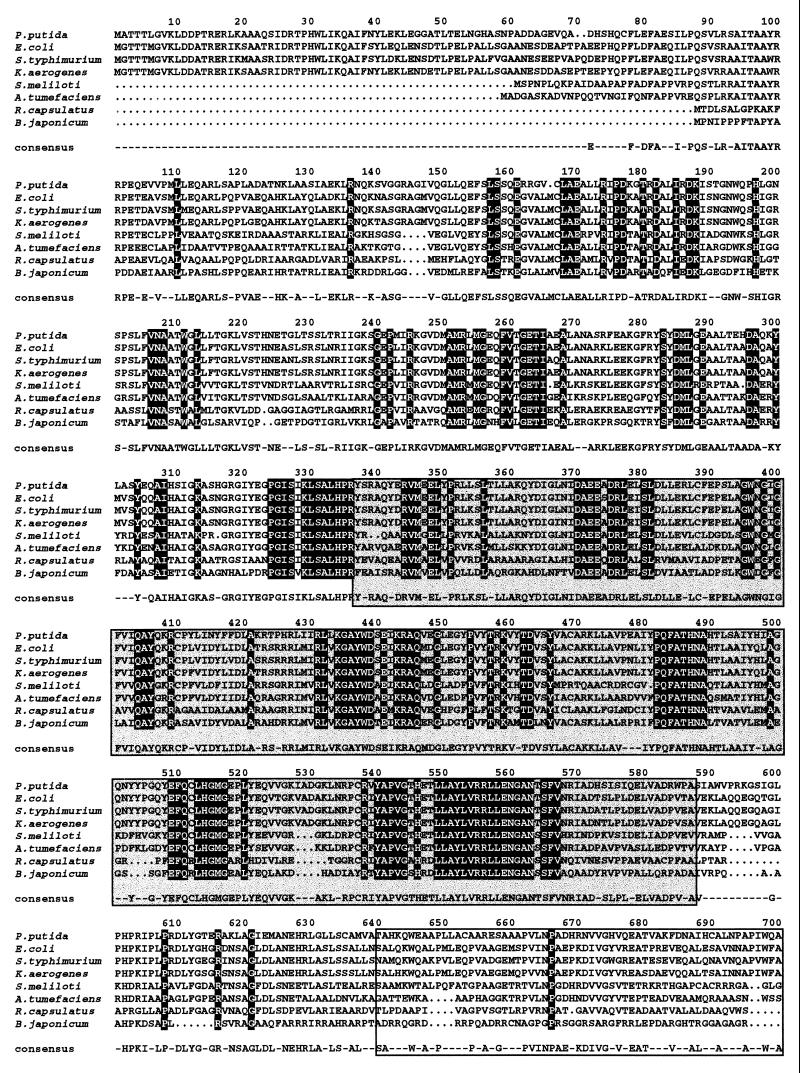
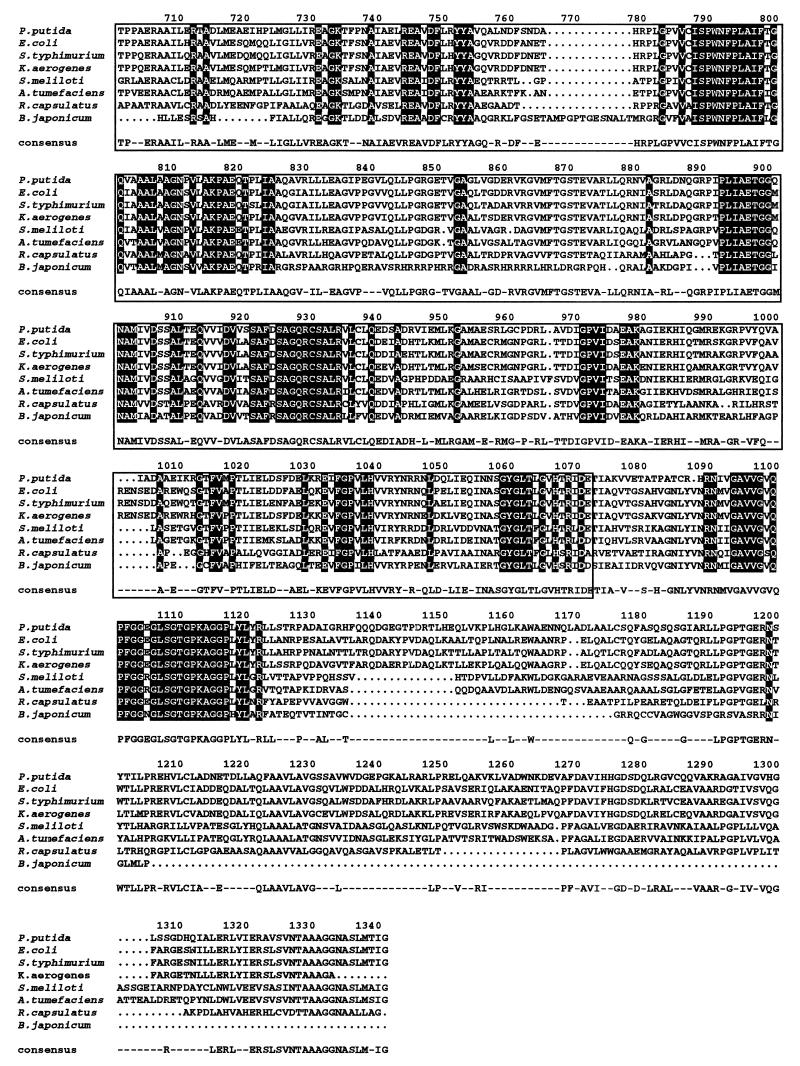
FIG. 3.
Sequence alignment of PutA proteins of prokaryotic origin. The strains and sources of the protein sequences were as follows: P. putida (this study); E. coli (31); Salmonella serovar Typhimurium (2); K. aerogenses (54); Agrobacterium tumefaciens (14); R. capsulatus (27); S. meliloti (25); B. japonicum (53). The ALIGN program was used. If the residue is identical in all the aligned proteins, it appears printed on a black background. If the residue is identical in 50% of the aligned proteins, it appears on a gray background. The amino acid chosen for the consensus was present at the given position in at least 50% of the aligned sequences. The PDH domain, residues 337 to 588, is shown in a grey box, and the P5CDH domain, residues 641 to 1074, is also boxed.