FIGURE 3.
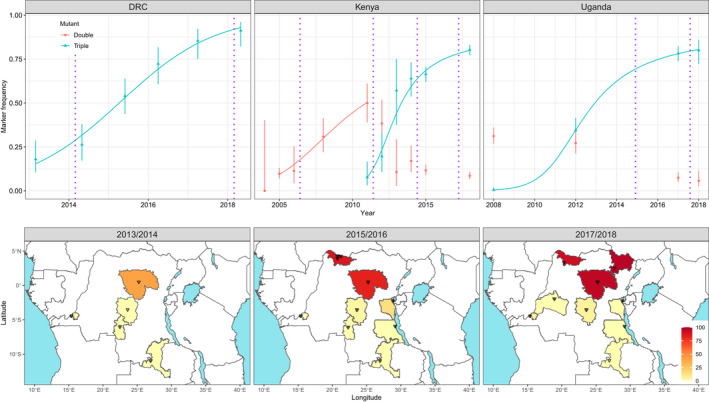
Observed and modelled changes in Cyp6aa/Cyp6p haplotype frequencies over time in Anopheles gambiae s.s. populations from DRC, Kenya and Uganda. Upper panel shows mutation frequency estimates derived from wild‐caught individuals from Kabondo, DRC; western Kenya and eastern Uganda, the 95% CIs for each observed data point were calculated according to Newcombe (1998). The model data, illustrated with blue or red curves, were generated from simultaneous maximum likelihood estimates of initial frequency and selection and dominance coefficients which were then used to parameterize standard recursive allele frequency change equation. The purple dotted lines indicate approximate timings of LLIN mass coverage campaigns in the sampled locations. The lower panel shows data from DRC tracking the emergence and spread of the triple mutant haplotype. Triangles indicate collection locations within each province and the scale bar indicates triple mutant frequency
