TABLE 5.
Localization frequencies of and complementation by GFP-FtsL mutants
| Construct | Structurea | Localization frequencyb
|
Complementatione
|
||
|---|---|---|---|---|---|
| No. of cells scored | Band at midcell (%) | Swap not fused to GFPc | Swap fused to GFPd | ||
| GFP-LLL | 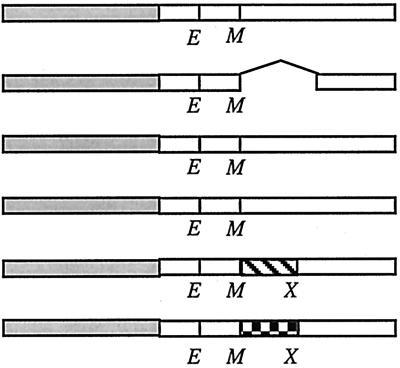 |
342 | 69 | +++ | +++ |
| GFP-LLΔL | 355 | − | ND | ||
| GFP-LLL(L70H) | 310 | 17 | + | ND | |
| GFP-LLL(L84D) | 380 | 28 | + | ND | |
| GFP-LLGCN4L | 232 | <1 | − | ND | |
| GFP-LLC/EBPL | 218 | <1 | − | ND | |
Open boxes represent domains of E. coli FtsL (FtsLEcol). Shaded boxes denote GFP. Hatched boxes represent GCN4 leucine zipper. The checkered box represents C/EBP leucine zipper. E and M indicate borders of the transmembrane domain where restriction sites for EagI and MscI, respectively, have been introduced. X indicates the position of the XhoI site introduced to allow partial swapping inside the region encoding the periplasmic domain of FtsL. In LLL, the former changes a Lys to an Arg, and the latter changes an Ala to a Gly. The cytoplasmic and transmembrane domains of LLL contain 37 and 20 amino acids, respectively. The periplasmic domain of LLL contains 64 amino acids.
Strains used: GFP-LLL, JMG287; GFP-LLΔL, JMG299; GFP-LLL(L70H), JMG303; GFP-LLL(L84D), JMG304; GFP-LLGCN4L, JMG301; GFP-LLC/EBPL, JMG302.
Strains used: LLL, JMG344; LLΔL, JMG350; LLL(L70H), JMG355; LLL(L84D), JMG356; LLGCN4L, JMG353; LLC/EBPL, JMG352.
Strain used: GFP-LLL, JMG367. ND, not determined.
For complementation test, plus signs indicate colony size and minus signs indicate no growth.
