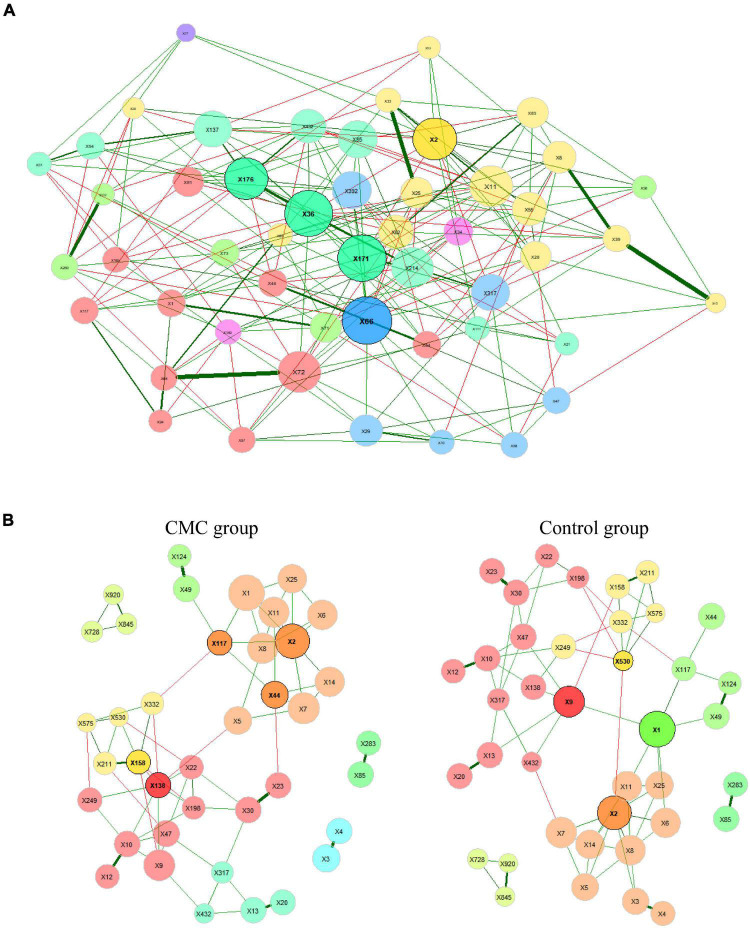
FIGURE 5.
Bacterial associations of samples in two groups. The SPRING method is used as an association measure. The estimated partial correlations are transformed into dissimilarities via the “signed” distance metric and the corresponding (non-negative) similarities are used as edge weights. Green edges correspond to positive estimated associations and red edges to negative ones. Eigenvector centrality is used for defining hubs (nodes with a centrality value above the empirical 95% quantile) and scaling node sizes. Hubs are highlighted by black borders. Node colors represent clusters, which are determined using greedy modularity optimization. The 100 most abundant taxa were analyzed in this part and (A) the complete association network where the 50 nodes with the highest degree are shown. (B) Comparison of bacterial associations in two groups. Centrality and clustering measures are adopted from the complete network. Species represented by the nodes are given in Supplementary Table 5.