Figure 3. General PRMT5 methylation targets and mechanisms.
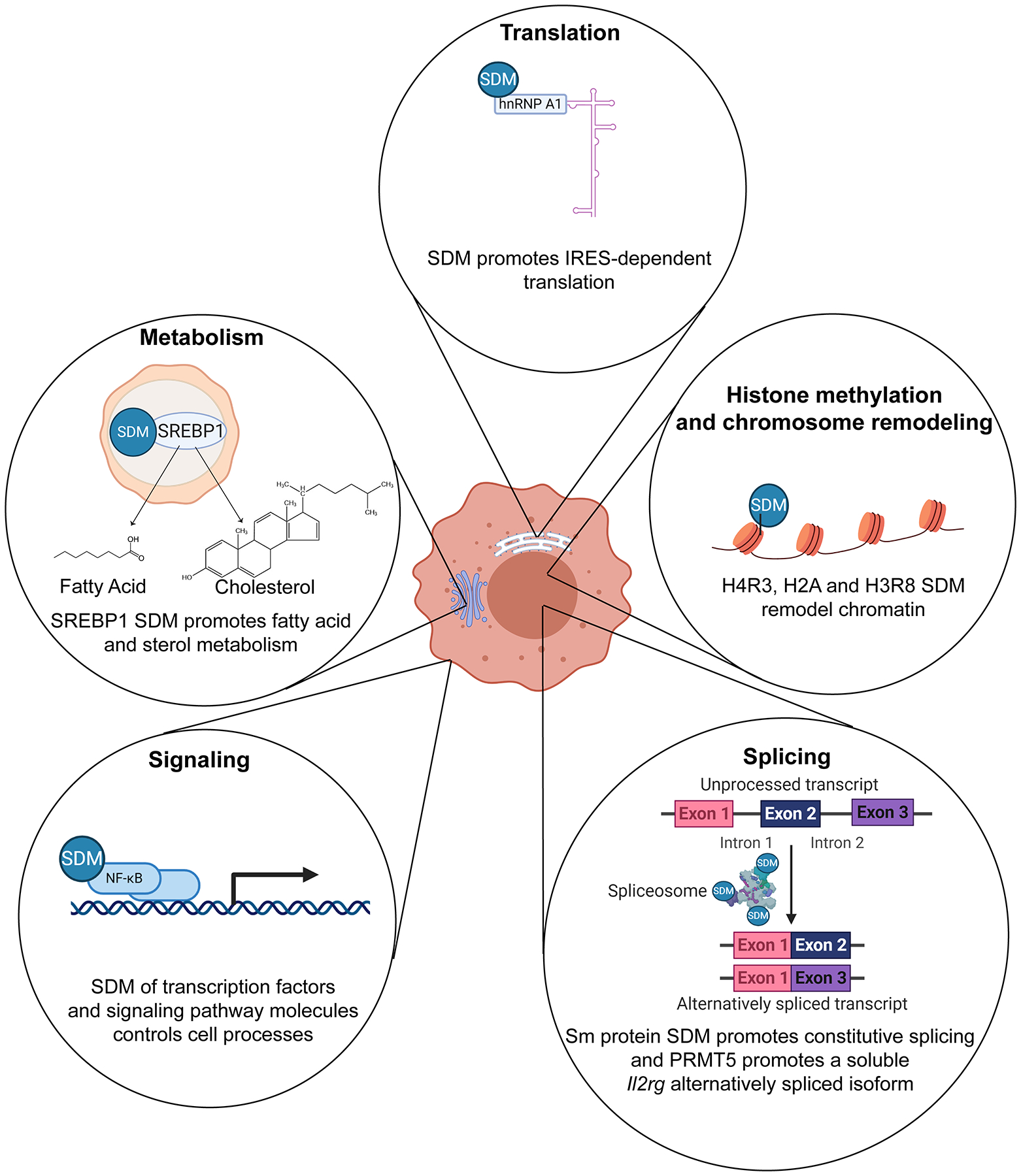
Studies in animal and plant cells show that PRMT5 symmetrically dimethylates (SDM) multiple targets in cytoplasmic and nuclear compartments, thereby influencing key cellular processes. Histone methylation: in the nucleus, SDM of histones H4R3, H3R8 and H2A generally results in chromatin repression via direct histone effects and recruitment of repressor complexes [8,89,90]. Most of the chromatin remodeling effects have been shown in non-T cell models [12]. Splicing: splicing proteins, such as SmD3 and SmB,B’, which can be found in the cytoplasm and nucleus, are also methylated by PRMT5 [82]. SDM of Sm proteins PRMT5 enhances spliceosome assembly and promotes alternative splicing switches [91]. SmD and SmB methylation has been observed in mouse T cells and is thought to increase Il2rg and Jak3 alternative splicing and expression [66]. Signaling: multiple cytoplasmic proteins, such as NF-kB and Vav-1, involved in receptor signaling pathways are methylated by PRMT5 [51,75]. Vav-1 methylation has been observed in Jurkat T cells and is believed to promote T cell costimulation [51]. Metabolism: SREBP is a lipid biogenesis transcription factor located in the endoplasmic reticulum and Golgi. Upon cleavage in the Golgi, SREBP is activated and translocates to the nucleus, where it transactivates fatty acid and cholesterol biosynthesis enzyme expression [92]. PRMT5 has been shown methylate SREBP and promote cholesterol metabolism in mouse T cells, thereby promoting Th17 differentiation [69]. Translation: PRMT5 has been shown to SDM ribonucleoprotein hnRNPA1 in cancer cells, thereby promoting translation of internal ribosomal entry site (IRES)-containing transcripts [78]. This figure was created using BioRender (https://biorender.com/).
