Abstract
Background
In Latin America, few studies have been carried out on methicillin-resistant Staphylococcus aureus carriage in the pediatric population. We conducted a survey of nasal S. aureus carriage in neonates and in children attending the pediatric outpatient clinics in a large Brazilian city with high antimicrobial consumption.
Methods
Pernasal swabs of neonates were collected upon admission and at discharge in four neonatal intensive care units and of children less than five years of age during outpatient visits. Methicillin-resistant S. aureus isolates were characterized for antibiotic susceptibility, mec gene presence, pulsed-field gel electrophoresis, spa type, SCCmec-type, multilocus sequence type, and presence of Panton-Valentine leukocidin genes.
Results
S. aureus was carried by 9.1% and 20.1% of the 701 neonates and of 2034 children attending the outpatient clinics, respectively; methicillin-resistant S. aureus carriage was detected in 0.6% and 0.2%, of the these populations, respectively. Healthcare-associated methicillin-resistant S. aureus strains found in neonates from neonatal intensive care units and outpatients were genetically related to the Brazilian (SCCmec-III, ST239) and to the Pediatric (SCCmec-IV, ST5) clones. Community-associated methicillin-resistant S. aureus was only detected in outpatients. None of the methicillin-resistant S. aureus strains contained the Panton-Valentine leukocidin gene. Methicillin-resistant S. aureus strains related to the Brazilian clone showed multidrug resistance pattern.
Conclusions
Despite the high antibiotic pressure in our area, and the cross transmission of the healthcare-associated methicillin-resistant S. aureus clones between neonatal intensive care units and outpatients, the prevalence of methicillin-resistant S. aureus carriage is still low in our setting.
Keywords: Staphylococcus aureus carriage, Methicillin-resistant Staphylococcus aureus, Neonates, Children
Introduction
Staphylococcus aureus is a common cause of community and healthcare-associated illness. Carriage of S. aureus is frequent in children and the anterior nares are the most consistent site for S. aureus colonization.1 Children colonized with methicillin-resistant S. aureus (MRSA) are potential reservoirs for the spread of MRSA in the community.2, 3 The relationship between MRSA nasal carriage and invasive staphylococcal infection has been well documented.4 A limited number of MRSA clonal lineages has been responsible for the majority of MRSA infections in several regions.
Since the emergence and rapid spread of community-associated MRSA (CA-MRSA) in previously healthy hosts,5 attention has been given to studies reporting changes in the epidemiology of MRSA colonization especially CA-MRSA in hospitalized patients6 and healthcare-associated MRSA (HA-MRSA) in healthy subjects.7, 8
In neonatal intensive care units (NICUs), neonates are at high risk for MRSA infections. Patients colonized by MRSA in NICUs present significant rates of bloodstream infection9 and are often transferred to other hospitals, contributing to the spread of MRSA in health facilities. Studies on MRSA carriage in neonates, however, are usually conducted under outbreak situations in NICUs.10, 11
Despite the severity and rapid global expansion of MRSA strains, the epidemiology of MRSA nasal carriage remains unclear. In Latin America, few molecular studies are available on MRSA carriage in the pediatric population.12, 13 To determine the extent the nasal carriage of MRSA is widespread in the pediatric population, we carried out a survey in neonates and in outpatient children in a large Brazilian city. We investigated molecular characteristics of MRSA isolates.
Materials and methods
Study area
The study was conducted in the municipality of Goiânia, capital of Goiás state, located in Central Brazil, a region with high rates of antibiotics consumption (Andrade 2012). In the 2010 census, the population of Goiânia was estimated at 1,302,001 inhabitants, with 20,014 live births, infant mortality at 14.6 deaths per 1000 live births, and 84,465 children under five years.14 Public health care is provided free of charge by the Brazilian Unified Health System, and an estimated 75% of the population use the public health system.15 In Goiania, children are first seen at the public healthcare centers (pediatric outpatient clinics) and referred to hospitalization whenever necessary. For this study, two pediatric populations were screened for MRSA, as described below.
Neonates admitted to NICUs
From June 2007 through November 2008 were screened for MRSA carriage all neonates admitted to four NICUs, which attend children from public and private health insurance. Taking together these NICUs comprise the majority of beds (n = 69) for neonates in the city. Data were collected on demographics (gender, date of birth, and age at NICU admission), each neonate's clinical history (length of hospital stay, congenital malformation, birth weight, preterm, timing of swab collection, nasogastric intubation, and diagnoses upon admission and at discharge), and variables related to their mothers (type of delivery, schooling, number of prenatal visits, and placental abruption).
Outpatient children
The carriage survey was conducted from July 2007 through July 2008 in children less than five years of age attending the major outpatient clinic of the city, served by the Brazilian public health insurance (Brazilian Unified Health System). Data were collected on child (gender, age, congenital malformation, skin and soft-tissue infections, hospitalization in the last six months, antibiotic use in the last three months, day care attendance in the last six months, acute otitis media in the last 12 months, pneumonia in the last 12 months, and tonsillitis in the last 12 months) and the family (mother's schooling, smoking at home, number of family members, and health worker in the family). We calculated that a sample size of 2000 children would be necessary to ascertain MRSA prevalence assuming a 1.2% prevalence of MRSA nasal carriers16 with a 95% confidence interval (95% CI), a precision of 0.7%, design effect of 1.8, and a refusal rate of 10%. The number of children who were recruited at the outpatient clinics per day took into account the laboratory capability for processing nasal specimens (approximately 10 swabs/day).
Collection of nasal specimens
Pernasal flocked swabs were collected by rotating the swab twice in the vestibule of the anterior nostrils of each child. In the outpatients, a single sample was collected from each child. At the NICUs, two samples were collected per child; the first collected upon admission and the second at discharge or death. After collection, the swabs were placed in a liquid medium (ESwab, Copan Diagnostics, Inc., Murrieta, CA, USA) and sent to the Microbiology Laboratory of the Federal University of Goiás for immediate processing.
Study ethics
The Institutional Review Board approval was given by the Ethics Committee of the Federal University of Goiás (# 008/2007). All parents or legal guardians signed copies of the informed consent form before recruitment.
Microbiological techniques
The specimens were plated onto mannitol salt agar (Difco® Laboratories Inc., Detroit, MI, USA) for S. aureus isolation. Trypticase soy agar plates, which were supplemented with 4% NaCl and oxacillin at 6 μg/mL, were used to screen MRSA isolates. After 24–48 h of incubation at 37 °C, the colonies that were suspected of being S. aureus were confirmed by Gram staining, catalase, coagulase, and PCR amplification of the femB gene (Jonas 1999).
Antimicrobial susceptibility testing
Susceptibility tests were determined by disk diffusion (Oxoid Ltd., Basingstoke, Hampshire, England) for oxacillin, cefoxitin, clindamycin, ciprofloxacin, erythromycin, rifampicin, tetracycline, trimethoprim–sulfamethoxazole, linezolid, and quinupristin/dalfopristin according to the Clinical Laboratory Standards Institute guideline.17 Isolates resistant to cefoxitin and oxacillin were submitted to the Etest® (AB Biodisk, Slona, Sweden) to determine the sensitivity to vancomycin. S. aureus strains ATCC 25923 and ATCC 29213 were used as quality controls.
MRSA definition
Isolates showing inhibition zones of ≤10 mm and ≤21 mm around 1 mg oxacillin (Oxoid, Cambridge, UK) and 30 μg cefoxitin (Oxoid, Cambridge, UK) disks, respectively, and that were positive for mecA gene by PCR, were characterized as MRSA.
Molecular typing
Detection of mecA gene and SCCmec types were investigated by multiplex PCR18 using COL (SCCmec type I), N315 (SCCmec type II), ANS46 (SCCmec type III), and MW2 (SCCmec type IV) as positive controls. Multilocus sequence typing was carried out as previously reported19 and submitted to the international database (http://www.mlst.net) to assign the sequence type (ST). Sequencing of the polymorphic X region of the protein A gene (spa) and the Panton-Valentine leukocidin (PVL) gene were performed as described elsewhere.20, 21 The sequences were analyzed using the Ridom Staph Type software (Ridom, Germany) to determine the spa types. The following international MRSA clones were included as controls: ST5-II (New-York/Japan, USA100), ST239-III (Brazilian/Hungarian), ST5-VI (Pediatric, USA800), and ST22-IV (EMRSA-15). The clonal relatedness was investigated by pulsed-field gel electrophoresis (PFGE) with SmaI macrorestriction according to standard procedures.22 The PFGE patterns were interpreted according to the criteria proposed by Tenover et al.,23 and the resulting dendrogram was constructed using Dice coefficients with BioNumerics software (version 5.0). Isolates were defined as epidemiologically related if they shared ≥80% similarity on the dendrogram.
Data analysis
Prevalence of MRSA and the corresponding 95% confidence interval (95% CI) were calculated. Multidrug resistance (MDR) was defined for isolates with resistance to three classes of antimicrobials and to β-lactams. Moving-average was used to calculate the percentage of colonization by S. aureus; the value attributed to a given month was the average colonization in that month, and the ones of the months prior and after the given month. Differences in lengths of stay in NICUs between neonates colonized and non-colonized by MRSA strains were evaluated by the Mann–Whitney test.
Results
Neonates
During the study period, a total of 710 neonates were admitted to the four NICUs, and nasal swabs were obtained from 701 (98.7%) of them. The characteristics of the neonates and their mothers are described in Table 1.The overall prevalence of S. aureus and MRSA was 9.1% (95% CI 7.2–11.4) and 0.6% (95% CI 0.2–1.4), respectively, and varied among the NICUs (Table 2). Four neonates carried MRSA strains; three of them acquired MRSA during the hospitalization period and had higher lengths of stay (median = 20 days) compared to neonates who were non-colonized by MRSA (median = 10 days) (p = 0.072).
Table 1.
Characteristics of children screened for Staphylococcus aureus and MRSAa nasal colonization. Goiânia, Brazil, 2007–2008.
| Variables | n/total | (%) |
|---|---|---|
| Neonatal intensive care units (n = 710)b | ||
| Male | 409/709 | (57.6) |
| Age at admission < 24 h | 519/701 | (74.0) |
| Low birthweight | 393/704 | (56.5) |
| Mother's schooling ≤ 8 years | 260/680 | (36.6) |
| Congenital malformation | 117/702 | (16.5) |
| Frequency of prenatal visits ≤ 6 | 418/663 | (58.9) |
| Cesarean section | 438/704 | (61.7) |
| Premature birth | 454/687 | (63.9) |
| Intubation | 374/690 | (52.7) |
| Nasogastric tube | 257/688 | (36.2) |
| Emergency department (n = 2034) | ||
| Males | 1108/2034 | (54.5) |
| Age ≥ 24 months | 784/2034 | (38.5) |
| Congenital malformation | 69/2031 | (3.4) |
| Skin and soft-tissue infections in the last 12 months | 570/2033 | (28.0) |
| Acute otitis in the last 12 months | 752/2034 | (37.0) |
| Pneumonia in the last 12 months | 623/2033 | (30.6) |
| Tonsillitis in the last 12 months | 1139/2033 | (56.0) |
| Hospitalization in the last 6 months | 586/2031 | (28.8) |
| Antibiotic use in the last 3 months | 948/1821 | (46.6) |
| Day care attendance in the last 6 months | 388/2033 | (19.1) |
| Mother's schooling ≤ 8 years | 893/2028 | (43.9) |
| Smoking at home | 695/2033 | (34.2) |
| Health worker in the family | 109/2034 | (5.4) |
| Number of family members > 4 | 676/2031 | (33.2) |
Methicillin-resistant Staphylococcus aureus.
The total does not always equals to 710.
Table 2.
Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus carriage according to the neonatal intensive care unit.
| Neonatal intensive care units |
S. aureus |
MRSA |
||||
|---|---|---|---|---|---|---|
| n | % | (95% CI) | n | % | (95% CI) | |
| #1 (n = 93) | 5 | 5.4 | (2.0–11.5) | 0 | 0.0 | – |
| #2 (n = 80) | 2 | 2.5 | (0.4–8.0) | 0 | 0.0 | – |
| #3 (n = 426) | 41 | 9.6 | (7.1–12.7) | 2 | 0.5 | (0.1–1.5) |
| #4 (n = 102) | 16 | 15.7 | (9.6–23.7) | 2 | 2.0 | (0.3–6.3) |
| Total (n = 701) | 64 | 9.1 | (7.2–11.4) | 4 | 0.6 | (0.3–1.6) |
Children attending the outpatients
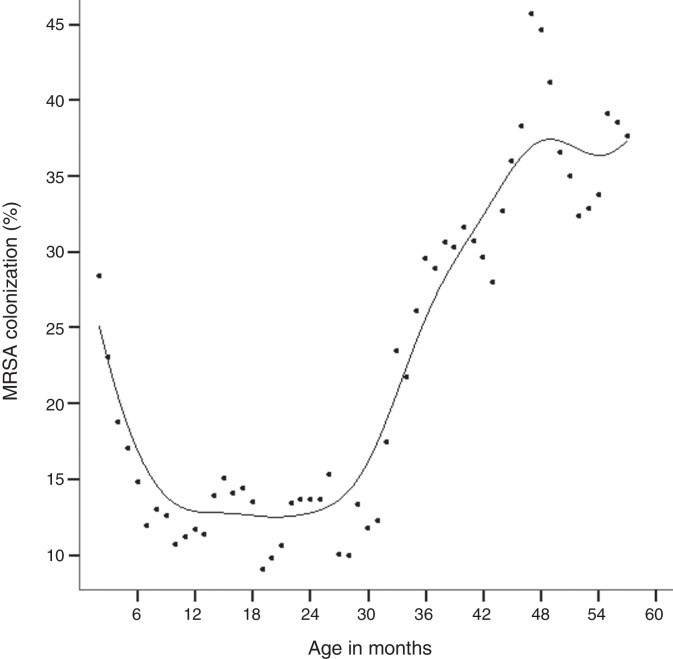
A total of 2034 children were enrolled for nasal swabbing during the study period. The mean age of the children was 21.6 months (SD = 16.0), and 31.8% of them had less than 12 months of age. The main clinical features of the participant children are shown in Table 1. Four hundred and eight children (20.1%; 95% CI: 18.4–21.9%) were colonized by S. aureus. Colonization by S. aureus showed an age-related distribution, with peak prevalence at 47 months of age (Fig. 1). Four children (0.2%; 95% CI: 0.1–0.5%) carried MRSA strains.
Fig. 1.
Age-related prevalence of Staphylococcus aureus colonization by age. Each dot represents a child colonized by S. aureus. Vertical arrows indicate the age of child colonized by the corresponding HA-MRSA and CA-MRSA clones.
Antimicrobial resistance of MRSA isolates
We identified 16 MRSA isolates from eight children. Six children had more than one isolate. MDR MRSA isolates were found in three children. Eleven MRSA isolates did not meet criteria for MDR, but they were resistant to different classes of antibiotics. MRSA isolates showed resistance to erythromycin (43.8%), clindamycin (37.5%), ciprofloxacin (25.0%) and trimethoprim–sulfamethoxazole (18.8%). All MRSA strains were susceptible to vancomycin.
MRSA molecular findings
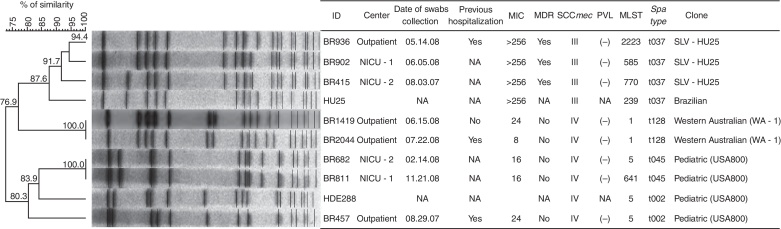
The majority of the MRSA strains clustered into two genomic groups; within each group we identified children from both, NICUs and outpatient clinics carrying MRSA strains genetically related to classical pandemic HA-MRSA Brazilian (SCCmec-III, ST239) and Pediatric (SCCmec-IV, ST5) clones (Fig. 2). CA-MRSA strains were only detected in outpatient children and were clonally related to Western Australian 1 clone/WA-1 (SCCmec-IV, ST128). Only the HA-MRSA Brazilian clone showed MDR pattern. None of the MRSA strains contained the PVL gene.
Fig. 2.
Molecular characteristics of the 8 MRSA strains from neonates admitted to neonatal intensive care units and children who visited the outpatient clinics. HDE288, representative Pediatric clone; HU25, representative Brazilian clone; NICU-1, neonatal intensive care unit of the Santa Bárbara Hospital; NICU-2, neonatal intensive care unit of the Hospital da Criança; NA, does not apply; MIC, minimum inhibitory concentration; MDR, multidrug-resistant; SCCmec, staphylococcal cassette chromosome mec; PVL, Panton-Valentine leukocidin; MLST, multilocus sequence type; SLV, single locus variant.
Discussion
Despite evidence that the MRSA incidence is increasing worldwide, studies on MRSA carriage in Latin America are still scarce. This study provides information on S. aureus and MRSA nasal carriage in outpatient children and in neonates admitted to NICUs in an urban area of Brazil with high antimicrobial pressure and infection control lapses.
We found that the frequency of S. aureus by age in children attending outpatients resembled a “J” curve, with higher prevalence in infants less than six months of age and in children older than 30 months of age. This result is consistent with studies by Bogaert et al.,24 which reported similar shape of age distribution of S. aureus carriage in healthy children.
The rates of MRSA carriage in NICUs herein were lower compared to studies from the US, Israel, and Taiwan.9, 11, 25 One possible explanation for the low MRSA carriage rate is that the present investigation was performed out of an outbreak, while the majority of studies in NICUs are usually conducted as a result of an outbreak of MRSA infection.
We also found lower prevalence of MRSA nasal colonization in outpatients when comparing to reports in children attending well-child health care services in the US3 and in Taiwan.26 Our findings, however, were comparable to those obtained in Southern Israel, where the prevalence of MRSA carriage in healthy infants aged 2–12 months ranged from 0.0% to 0.7%.27 Previous investigation in Brazil conducted in outpatient children16 detected a higher prevalence of MRSA nasopharyngeal carriage (1.0%) than that found in the current study (0.2%), possibly because in the previous study the nasopharyngeal specimens were obtained during the winter period, when bacterial colonization of respiratory tract is expected to be higher compared to colonization rates obtained from non-seasonal periods. Nevertheless, comparison of our results on MRSA carriage with other studies is not straightforward due to differences in study designs, inclusion criteria, epidemiological scenarios, and age groups.
Despite the low colonization rates by MRSA found herein, this might turn out to be a serious concern. We identified three pandemic clones of MRSA (Brazilian, Pediatric, and WA1). The CA-MRSA WA-1 clone, detected in outpatients, has been identified in patients with bloodstream infections from South and Southeast regions of Brazil,28 indicating the dissemination of this clone across the country. The HA-MRSA Brazilian clone was detected in an outpatient child. This clone has been disseminated through hospitals in many countries of Latin America.29 Previously, we described the presence of the Brazilian clone in healthy children.16 In all these cases, the children had been admitted to hospitalization in the previous six months.
One interesting finding of this study is the presence of the HA-MRSA Pediatric clone recovered from outpatient children. This clone has been identified in hospitals of the Southeast and Northeast Regions of Brazil30, 31 but this is the first detection of the Pediatric clone in Central Brazil, suggesting that this clone is settling in Brazilian hospitals. This observation confirms that the Pediatric clone is spread in the community, increasing the chances of expanding its reservoir.
Some limitations of our study should be mentioned. First, as we collected nasal swabs of neonates only at admittance and discharge, we were not able to evaluate children who were intermittently colonized by MRSA in NICUs. Second, the small number of MRSA isolates resulted in an insufficient precision of the prevalence rates of MRSA carriage. Third, in outpatients, we did not enroll children aged higher than five years of age, as the peak of MRSA carriage is expected to be at 11 years of age.24
In conclusion, we found low rates of MRSA carriage among children in an area with high antimicrobial pressure. However, we detected CA-MRSA, and HA-MRSA clones in the community, which could contribute for an invisible MRSA reservoir, requiring a close monitoring of the MRSA epidemiology in both community and healthcare environment.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by the Brazilian National Council for Scientific and Technological Development/CNPq. We are grateful to all pediatricians at the participant hospitals. We would also like to thank Copan Diagnostics Inc. for swab donations. A.L.A. (Grants 482646/2007-1; 306096/2010-2) and A.K. (Grant 301199/2009-8) are research fellows of CNPq.
References
- 1.Wertheim H.F., Melles D.C., Vos M.C., et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 2.Chen C.J., Hsu K.H., Lin T.Y., et al. Factors associated with nasal colonization of methicillin-resistant Staphylococcus aureus among healthy children in Taiwan. J Clin Microbiol. 2011;49:131–137. doi: 10.1128/JCM.01774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creech C.B., 2nd, Kernodle D.S., Alsentzer A., Wilson C., Edwards K.M. Increasing rates of nasal carriage of methicillin-resistant Staphylococcus aureus in healthy children. Pediatr Infect Dis J. 2005;24:617–621. doi: 10.1097/01.inf.0000168746.62226.a4. [DOI] [PubMed] [Google Scholar]
- 4.Lo W.T., Lin W.J., Tseng M.H., et al. Methicillin-resistant Staphylococcus aureus in children, Taiwan. Emerg Infect Dis. 2006;12:1267–1270. doi: 10.3201/eid1208.051570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David M.Z., Daum R.S. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey A.J., Della-Latta P., Huard R., et al. Changes in the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2010;31:613–619. doi: 10.1086/652526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho P.L., Chiu S.S., Chan M.Y., et al. Molecular epidemiology and nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus among young children attending day care centers and kindergartens in Hong Kong. J Infect. 2012;64:500–506. doi: 10.1016/j.jinf.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Vasoo S., Singh K., Chow C., et al. Health care-associated methicillin-resistant Staphylococcus aureus colonization in children attending day care centers in Singapore. Pediatr Infect Dis J. 2012;31:213–214. doi: 10.1097/INF.0b013e318243e209. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y.C., Chou Y.H., Su L.H., Lien R.I., Lin T.Y. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics. 2006;118:469–474. doi: 10.1542/peds.2006-0254. [DOI] [PubMed] [Google Scholar]
- 10.Sax H., Posfay-Barbe K., Harbarth S., et al. Control of a cluster of community-associated, methicillin-resistant Staphylococcus aureus in neonatology. J Hosp Infect. 2006;63:93–100. doi: 10.1016/j.jhin.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Regev-Yochay G., Rubinstein E., Barzilai A., et al. Methicillin-resistant Staphylococcus aureus in neonatal intensive care unit. Emerg Infect Dis. 2005;11:453–456. doi: 10.3201/eid1103.040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartoloni A., Pallecchi L., Fernandez C., et al. Low prevalence of methicillin-resistant Staphylococcus aureus nasal carriage in urban and rural community settings in Bolivia and Peru. Int J Infect Dis. 2012 doi: 10.1016/j.ijid.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Gardella N., Murzicato S., Di Gregorio S., et al. Prevalence and characterization of methicillin-resistant Staphylococcus aureus among healthy children in a city of Argentina. Infect Genet Evol. 2011;11:1066–1071. doi: 10.1016/j.meegid.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 14.DATASUS; Brasília, DF: 2010. Brazilian Ministry of Health. Informações de Saúde.http://www2.datasus.gov.br/DATASUS/index.php?area=02 Available at: [accessed 01.07.12] [Google Scholar]
- 15.Paim J., Travassos C., Almeida C., Bahia L., Macinko J. The Brazilian health system: history, advances, and challenges. Lancet. 2011;377:1778–1797. doi: 10.1016/S0140-6736(11)60054-8. [DOI] [PubMed] [Google Scholar]
- 16.Lamaro-Cardoso J., Castanheira M., de Oliveira R.M., et al. Carriage of methicillin-resistant Staphylococcus aureus in children in Brazil. Diagn Microbiol Infect Dis. 2007;57:467–470. doi: 10.1016/j.diagmicrobio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute . 10th ed. CLSI; Wayne, PA: 2009. Performance standards for antimicrobial susceptibility testing. Supplement M100-S19. [Google Scholar]
- 18.Milheiriço C., Oliveira D.C., de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 19.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koreen L., Ramaswamy S.V., Graviss E.A., et al. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lina G., Piemont Y., Godail-Gamot F., et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 22.Chung M., de Lencastre H., Matthews P., et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 23.Tenover F.C., Arbeit R.D., Goering R.V., et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogaert D., van Belkum A., Sluijter M., et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 25.Sarda V., Molloy A., Kadkol S., et al. Active surveillance for methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit. Infect Control Hosp Epidemiol. 2009;30:854–860. doi: 10.1086/605321. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y.C., Hwang K.P., Chen P.Y., Chen C.J., Lin T.Y. Prevalence of methicillin-resistant Staphylococcus aureus nasal colonization among Taiwanese children in 2005 and 2006. J Clin Microbiol. 2007;45:3992–3995. doi: 10.1128/JCM.01202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adler A., Givon-Lavi N., Moses A.E., Block C., Dagan R. Carriage of community-associated methicillin-resistant Staphylococcus aureus in a cohort of infants in southern Israel: risk factors and molecular features. J Clin Microbiol. 2010;48:531–538. doi: 10.1128/JCM.02290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro A., Coronado A.Z., Silva-Carvalho M.C., et al. Detection and characterization of international community-acquired infections by methicillin-resistant Staphylococcus aureus clones in Rio de Janeiro and Porto Alegre cities causing both community- and hospital-associated diseases. Diagn Microbiol Infect Dis. 2007;59:339–345. doi: 10.1016/j.diagmicrobio.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Noriega E., Seas C., Guzman-Blanco M., et al. Evolution of methicillin-resistant Staphylococcus aureus clones in Latin America. Int J Infect Dis. 2010;14:e560–e566. doi: 10.1016/j.ijid.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Sousa-Junior F.C., Silva-Carvalho M.C., Fernandes M.J., et al. Genotyping of methicillin-resistant Staphylococcus aureus isolates obtained in the Northeast region of Brazil. Braz J Med Biol Res. 2009;42:877–881. doi: 10.1590/s0100-879x2009005000018. [DOI] [PubMed] [Google Scholar]
- 31.de Miranda O.P., Silva-Carvalho M.C., Ribeiro A., et al. Emergence in Brazil of methicillin-resistant Staphylococcus aureus isolates carrying SCCmecIV that are related genetically to the USA800 clone. Clin Microbiol Infect. 2007;13:1165–1172. doi: 10.1111/j.1469-0691.2007.01830.x. [DOI] [PubMed] [Google Scholar]