Figure 1.
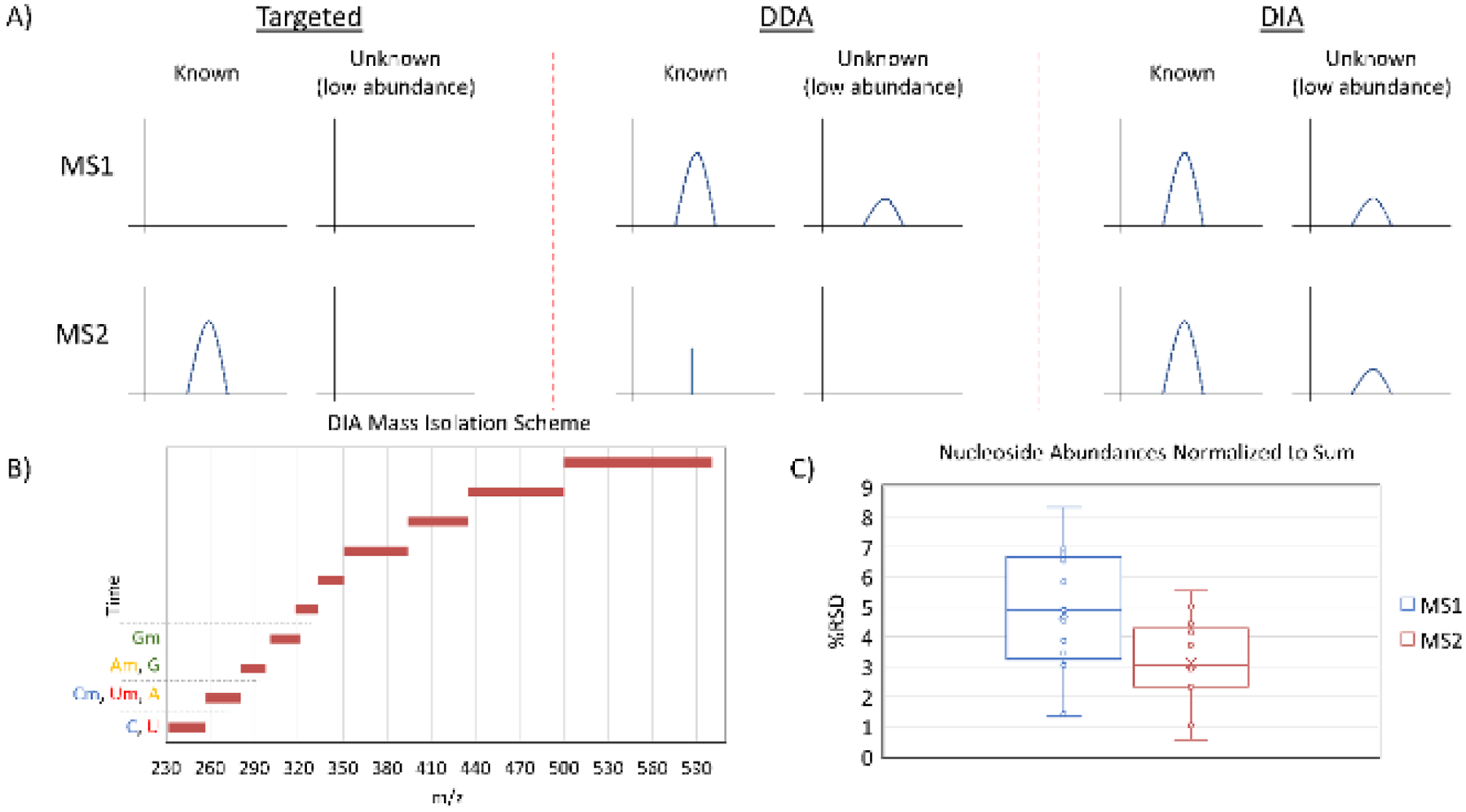
A) A comparison of MS acquisition approaches. Targeted acquisition is often performed using a QQQ mass spectrometer, which isolates a mass, fragments the molecule, and then isolates and detects a single fragment ion. DDA selects masses for fragmentation based on criteria such as abundance and often excludes previously fragmented masses from selection, resulting in few MS2 scans of abundant nucleosides and no MS2 scans of low abundance nucleosides. DIA fragments all masses in designated windows in each scan cycle, ensuring adequate points across a peak at the MS1 and MS2 levels for all nucleosides. B) The DIA mass isolation scheme designed in this work. The scan windows prevent different ribose modification states from producing the same fragment ion in the same MS2 scan from different precursors. C) A comparison of %RSD values for 5 HeLa biological replicates injected at different concentrations (1x to 5x). The nucleosides used were C, U, Ψ, A, G, m3C, m5C, Cm, I, m1A, m6A, Am, and m1acp3Ψ, and abundances were normalized to the sum of the included analytes. Within the same runs, MS2 was significantly more consistent than MS1 analysis (P < 0.05).
