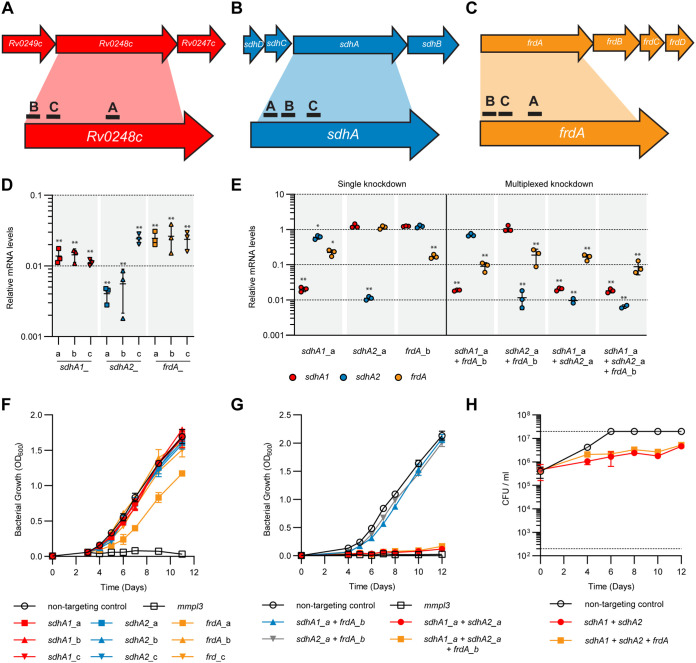
FIG 1.
Single and multiplexed transcriptional repression of sdhA1, sdhA2, and frdA in M. tuberculosis using CRISPRi. (A to C) Location of sgRNAs targeting the catalytic subunits of Sdh1 (A), Sdh2 (B), and Frd (C) in M. tuberculosis. sgRNAs were coexpressed with a dCas9sth1 under the control of an ATc-inducible promoter. (D) CRISPRi achieves high-level knockdown of target genes in single repression constructs. RNA was harvested 72 h after inducing knockdown (100 ng/mL ATc) and quantified by qPCR. mRNA is expressed relative to a strain expressing a nontargeting control. Results are mean ± standard deviation for three technical triplicates. ** indicates a P value of <0.005 from a one-way analysis of variance (ANOVA) with a Dunnett correction comparing each sgRNA to the nontargeting control. (E) CRISPRi achieves high-level knockdown of sdhA1, sdhA2, and frdA in single, double, and triple gene repression constructs. Gene knockdown was quantified and visualized as in panel D. Statistical significance was calculated using a one-way analysis of variance and a Dunnett test for multiple comparisons of the gene expression in each strain against the nontargeting control; *, P < 0.01; **, P < 0.001. (F) Consequences of the single knockdown of sdhA1, sdhA2, and frdA for the growth of M. tuberculosis in 7H9 medium supplemented with OADC. Knockdown was induced at time zero with ATc (100 ng/mL). An mmpL3-targeting sgRNA, which has bactericidal consequences for the viability of M. tuberculosis (48), was included as a positive inhibition control. The means and standard deviation for three replicates are shown. (G and H) Consequences of multiplexed sdhA1, sdhA2, and frdA gene repression for the growth (G) and viability (H) of M. tuberculosis in 7H9 medium supplemented with OADC. Dotted horizontal lines in panel H represent the upper and lower limits of detection of CFU. The means and standard deviation for three replicates are shown. Data in panels G and H are representative of three independent experiments.