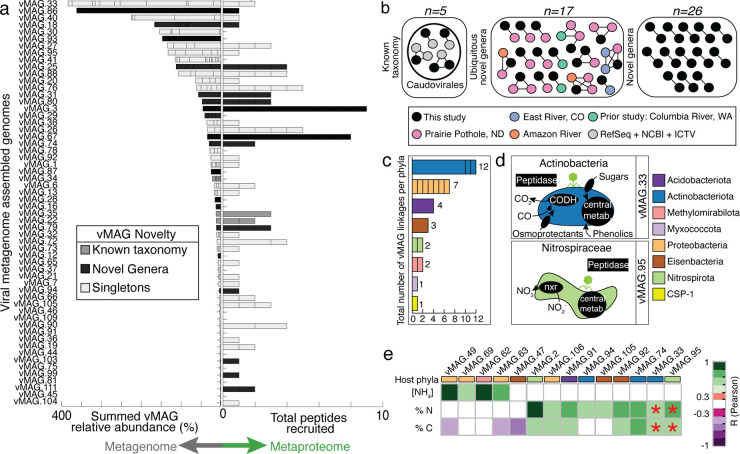
FIG 5.
Viruses in HUM-V are taxonomically novel, express genes, and may play roles in microbial host metabolism and river geochemistry. (a) The genomic relative abundance (left side) and total peptides recruited (right side) for each vMAG population. Bars are colored by clustering of vMAGs from this study with (i) viruses of known taxonomy (dark gray), (ii) novel genera (black), and (iii) no clustering with members from any database (light gray, singletons), as defined in Materials and Methods. (b) Similarity network of the few vMAGs from our study (denoted in black) that clustered to viruses with known taxonomy (gray) or to viruses in other freshwater, publicly available data sets (denoted in pink, purple, orange, and turquoise) and the remaining clusters of viruses that were novel (e.g., did not cluster with prior vMAGs). The full network table including singletons is shown in Table S4. (c) The total number of vMAGs (n = 32) with putative host linkages. Each bar represents a phylum, and lines within bars indicate the number of linkages for specific MAGs within that phylum. (d) Subset of cartoons of microbial metabolisms for two representative MAGs with putative viral linkages, with the genes shown in black text boxes denoting processes detected in proteomics. (e) Correlations between a subset of vMAGs, with rectangle colors denoting the putative host taxonomy. Correlations between these vMAGs and ecosystem geochemistry (concentration of NH4 [μg/g], %N, and %C) are reported, with significant correlation coefficients denoted by purple-green shading according to the legend. Red asterisks indicate that the vMAG relative abundance predicted a key environmental variable by sparse partial least-squares (sPLS) regression; note that these two vMAGs are shown in panel d.