Extended Data Figure 5: Localization-based sensitivity analysis.
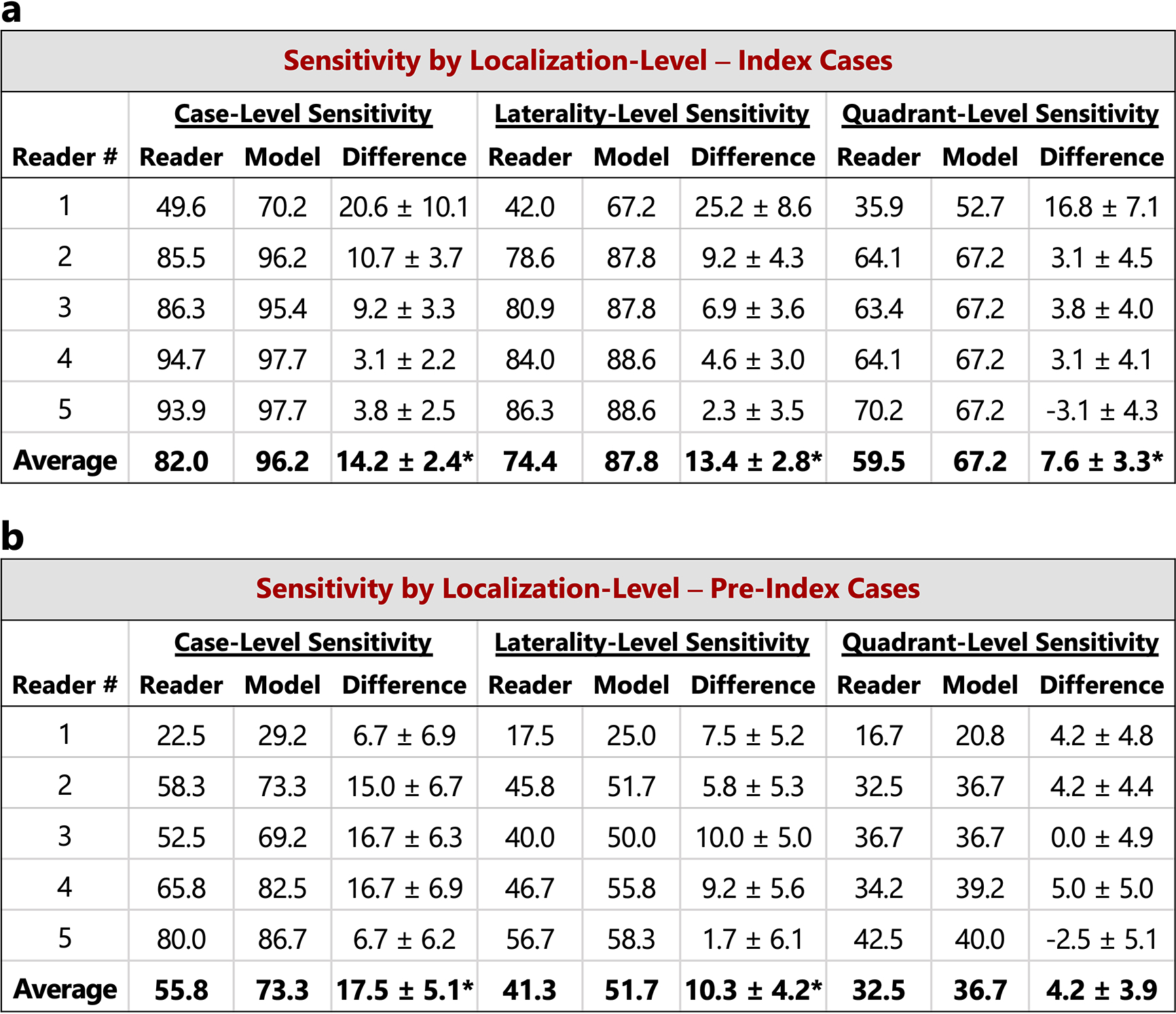
In the main text, case-level results are reported. Here, we additionally consider lesion localization when computing sensitivity for the reader study. Localization-based sensitivity is computed at two levels - laterality and quadrant (see Methods). As in Figure 2 in the main text, we report the model’s sensitivity at each reader’s specificity (96.1, 68.2, 69.5, 51.9, and 48.7 for Readers 1–5 respectively) and at the reader average specificity (66.9). a) Localization-based sensitivity for the index cases (131 cases). b) Localization-based sensitivity for the pre-index cases (120 cases). For reference, the case-level sensitivities are also provided. We find that the model outperforms the reader average for both localization levels and for both index and pre-index cases (*p<0.05; Specific p-values: index- laterality: p<1e–4, index - quadrant: p=0.01, pre-index - laterality: p=0.01, pre-index - quadrant: p=0.14). The results in the tables below correspond to restricting localization to the top scoring predicted lesion for both reader and model (see Methods). If we allow localization by any predicted lesion for readers while still restricting the model to only one predicted bounding box, the difference between the model and reader average performance is as follows (positive values indicate higher performance by model): index - laterality: 11.2±2.8 (p=0.0001), index - quadrant: 4.7±3.3 (p=0.08), pre-index - laterality: 7.8±4.2 (p=0.04), pre-index - quadrant: 2.3±3.9 (p=0.28). P-values and standard deviations were computed via bootstrapping. Finally, we note that while the localization-based sensitivities of the model may seem relatively low on the pre-index cases, the model is evaluated in a strict scenario of only allowing one box per study and crucially, all of the pre-index effectively represent “misses” in the clinic. Even when set to a specificity of 90% [36], the model still detects a meaningful number of the missed cancers while requiring localization, with a sensitivity of 37% and 28% for laterality and quadrant localization, respectively.
