Extended Data Figure 6: Reader study case characteristics and performance breakdown.
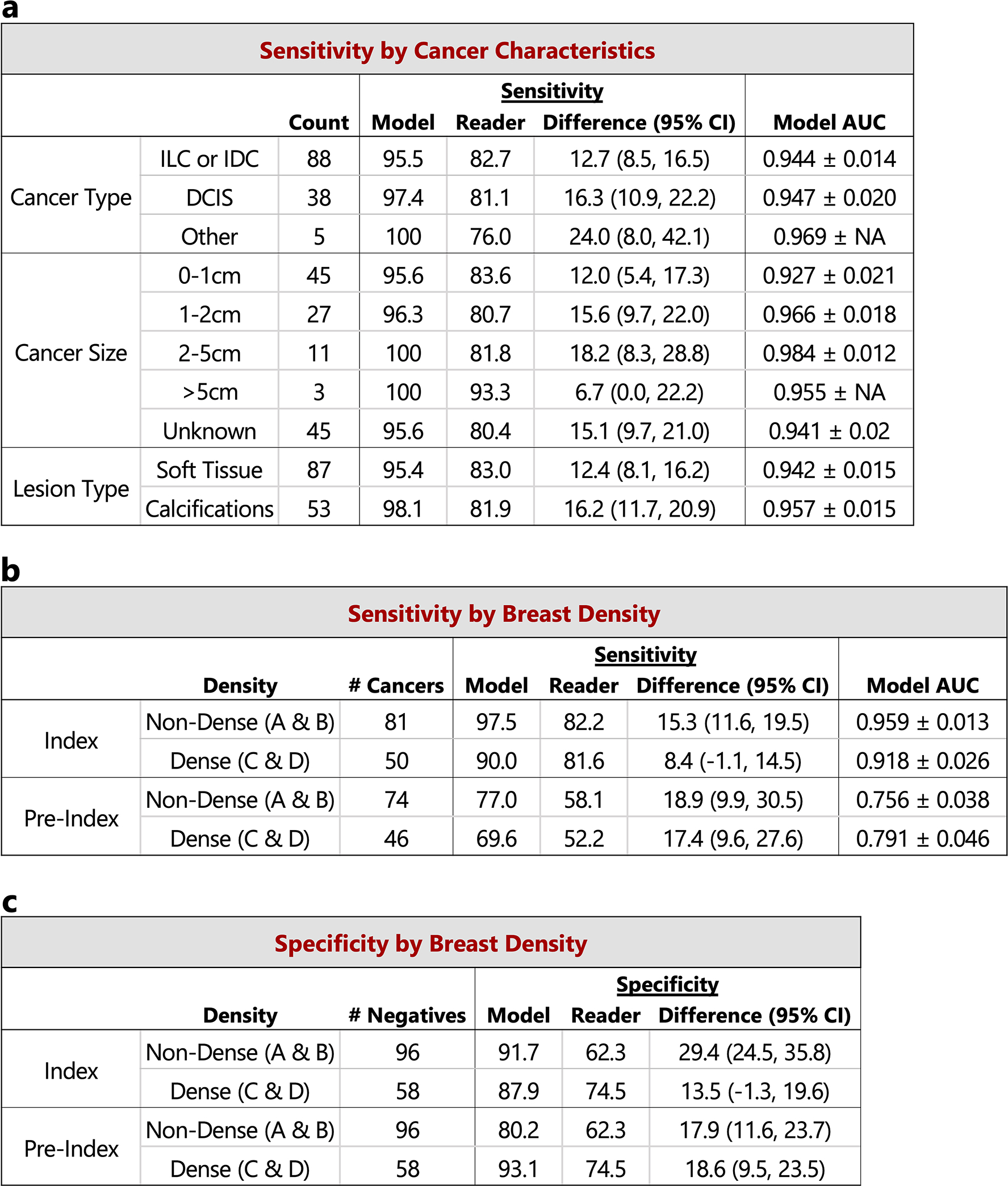
The performance of the proposed deep learning model compared to the reader average grouped by various case characteristics is shown. For sensitivity calculations, the score threshold for the model is chosen to match the reader average specificity. For specificity calculations, the score threshold for the model is chosen to match the reader average sensitivity. a) Sensitivity and model AUC grouped by cancer characteristics, including cancer type, cancer size, and lesion type. The cases correspond to the index exams since the status of these features are unknown at the time of the pre-index exams. Lesion types are grouped by soft tissue lesions (masses, asymmetries, and architectural distortions) and calcifications. Malignancies containing lesions of both types are included in both categories (9 total cases). ‘NA’ entries for model AUC standard deviation indicate that there were too few positive samples for bootstrap estimates. The 154 confirmed negatives in the reader study dataset were used for each AUC calculation. b) Sensitivity and model AUC by breast density. The breast density is obtained from the original radiology report for each case. c) Specificity by breast density. Confidence intervals and standard deviations were computed via bootstrapping.
