Figure 1. Primary structure of Schistocerca gregaria proteinase inhibitor 2 (SGPI-2).
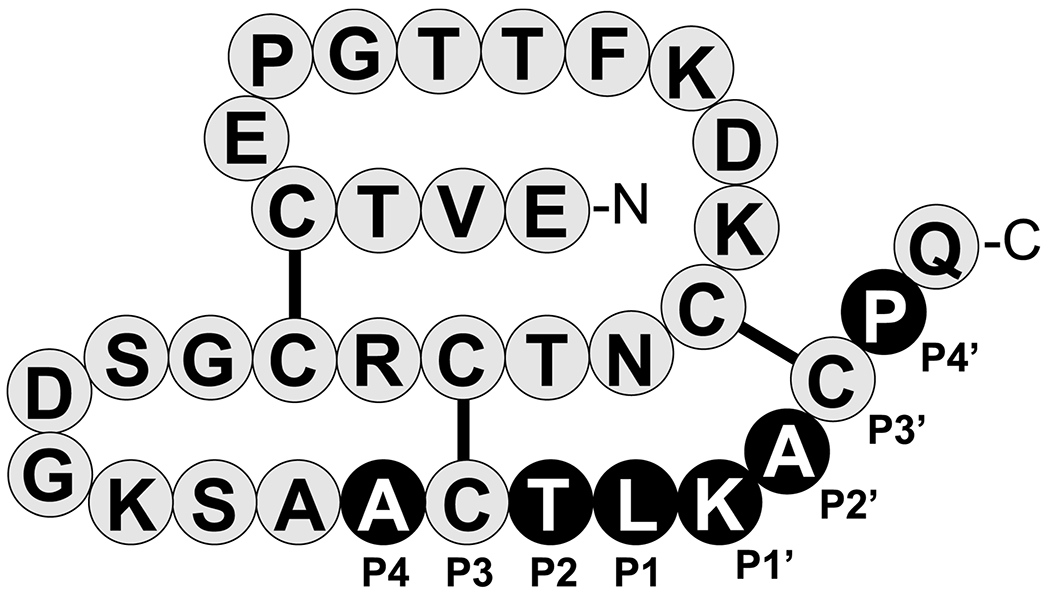
Canonical reactive loop residues are labeled P4-P4’ according to Schechter and Berger, where the reactive-site peptide-bond corresponds to P1-P1’ [6]. Residues that were fully randomized for creating the SGPI-2-phage library are highlighted as white letters on black background. The disulfide bridges are indicated. The figure was originally published in The Journal of Biological Chemistry: Szabó A.; Héja D.; Szakács D.; Zboray K.; Kékesi K. A.; Radisky E. S.; Sahin-Tóth M. and Pál G. (2011) High-affinity small protein inhibitors of human chymotrypsin C (CTRC) selected by phage display reveal unusual preference for P4’ acidic residues. J. Biol. Chem. 286 (25) pp. 22535-22545, Copyright 2011 by The American Society for Biochemistry and Molecular Biology [12].
