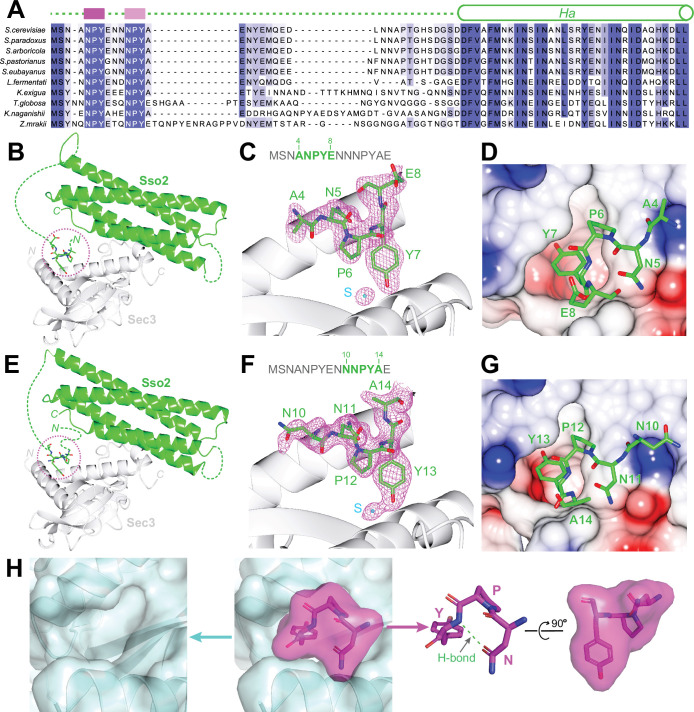
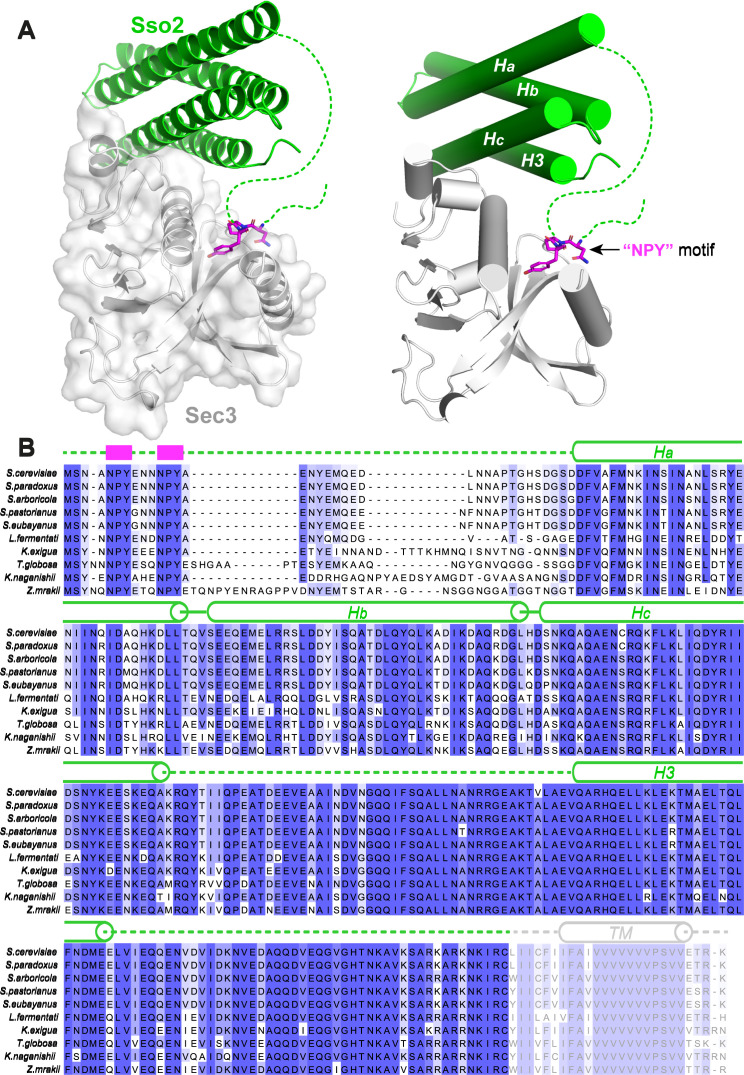
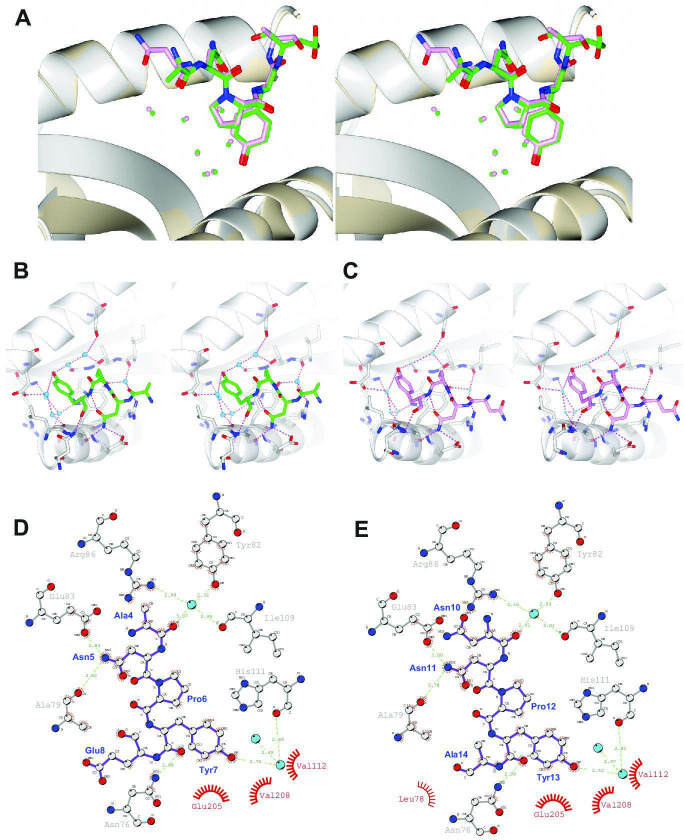
Figure 2. Structural analyses of the interaction between the NPY motifs of Sso2 and Sec3.
(A) Sequence alignments of Sso2 homologs. Conserved residues are shaded in dark (highly conserved) or light blue (partially conserved). The two NPY motifs are marked as magenta blocks above the aligned sequences, which are connected to helix Ha via a variable linker. (B) Ribbon diagram of the crystal structure of the Sso2/Sec3 complex with first NPY motif (shown as sticks) of Sso2 bound to Sec3. (C) An enlarged view of the NPY motif in (B) together with the 2Fo–Fc map contoured at 1.5σ. (D) Sticks of the NPY motif on top of an electrostatic surface plot of Sec3. (E–G) Crystal structure shows how the second NPY motif of Sso2 interacts with Sec3. (H) Separate views of the binding site show the cork-like NPY motif (magenta) and the complementary cradle-like pocket on Sec3 (light blue). The structure is shown as semitransparent surface together with ribbon diagrams (Sec3) or sticks (Sso2). The broad top part of the ‘cork’ of Sso2 is stabilized by a hydrogen bond between the carboxyl group of the Asn (N) sidechain and the amide proton of Tyr (Y).