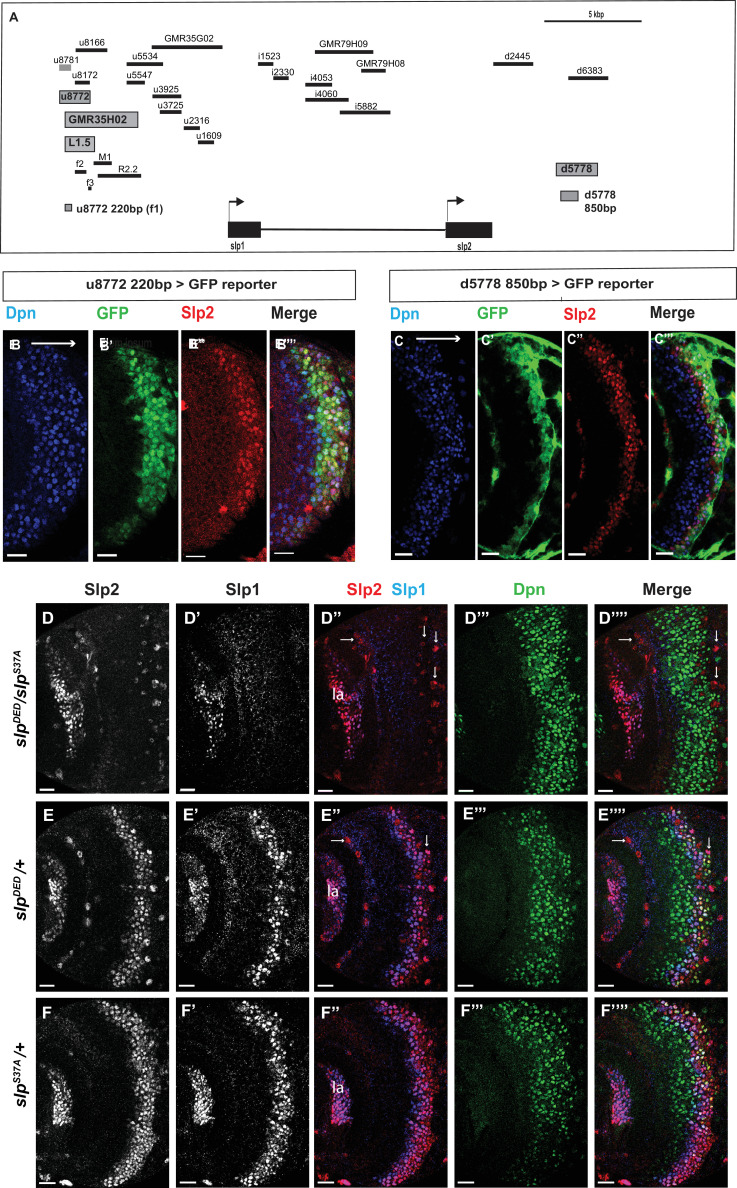
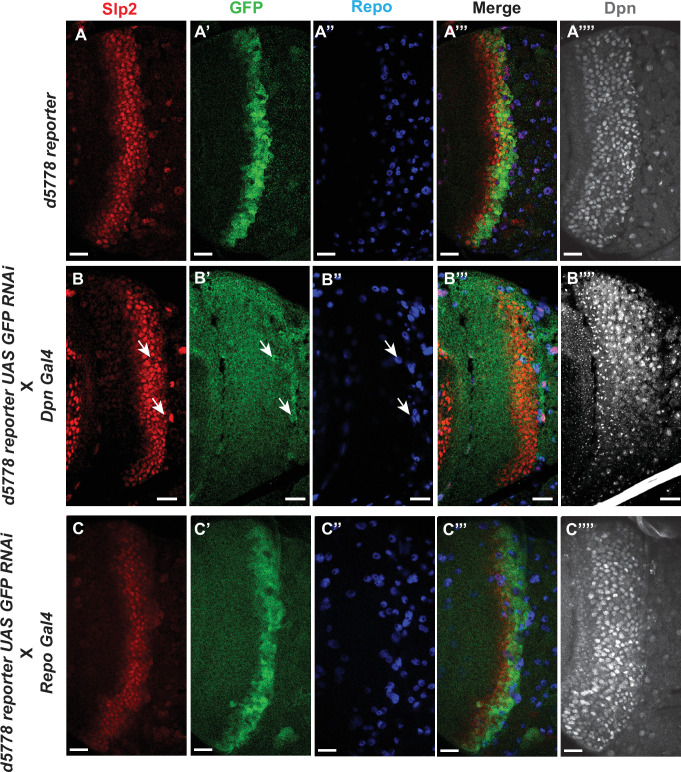
Figure 2. Transcription of slp1 and slp2 in medulla neuroblasts is regulated by two enhancers of lengths 220 bp and 850 bp, respectively.
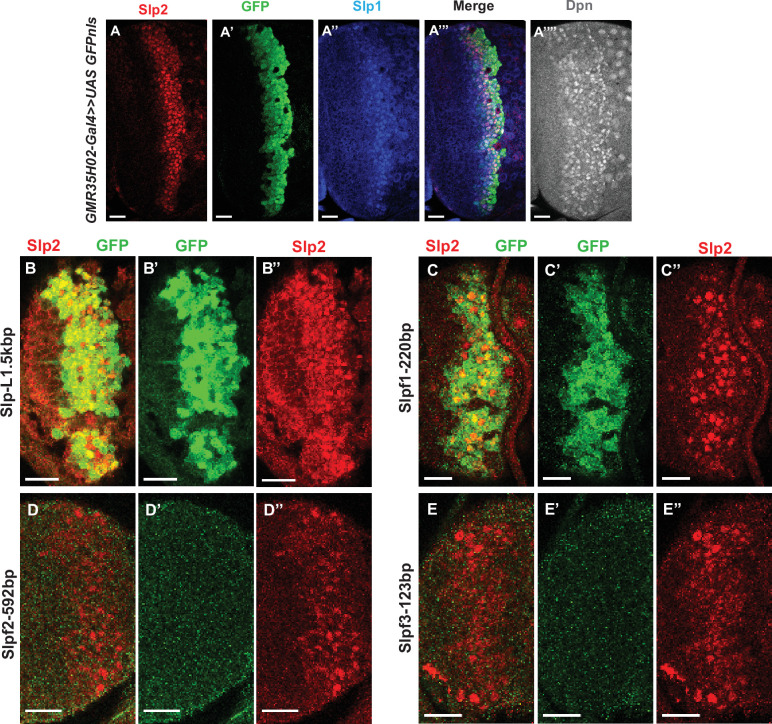
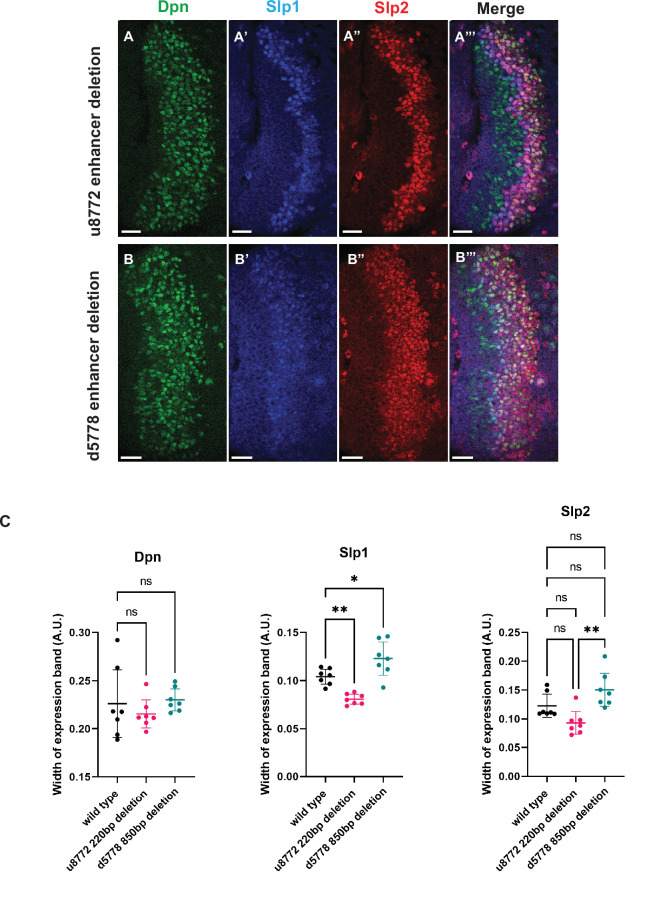
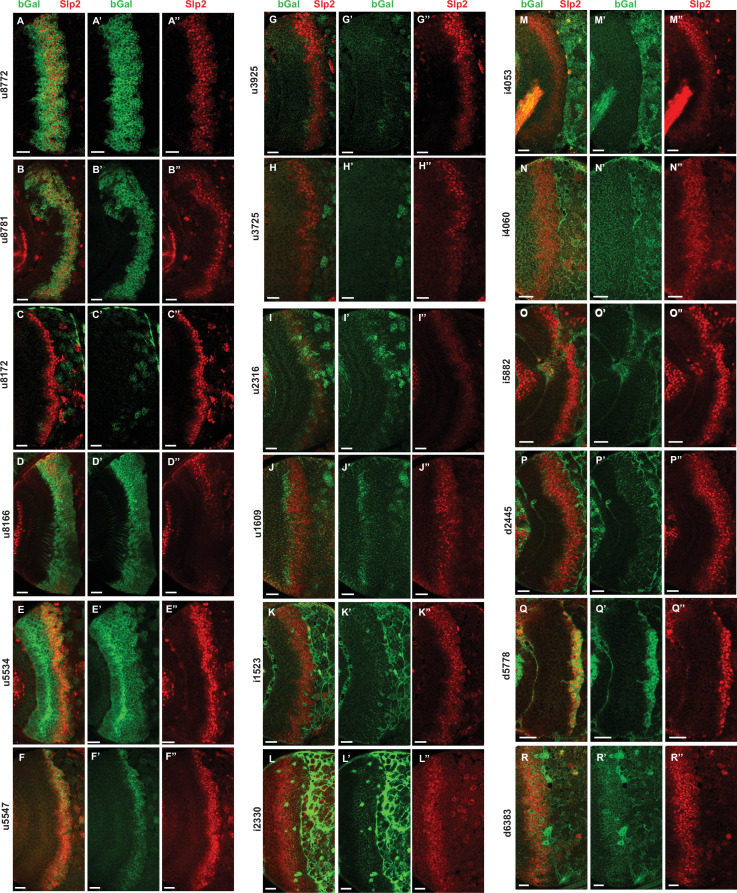
(A) A schematic of the enhancer screening. slp1 and slp2 genes and transcriptional enhancers identified as regulating their expression are shown as black and grey boxes, respectively. Positions of slp1 coding locus (2 L:3,825,675.3,827,099 [+]), slp2 coding locus (2 L:3,836,840.3,839,185 [+]), GMR35H02 (2 L:3,817,265.3,821,014) and the REDfly enhancers u8772 (2 L:3,816,967.3,818,532) and d5778 (2 L:3,842,530.3,844,660) are shown relative to one another. A 220 bp enhancer located within the genomic segment covered by GMR35H02 and an 850 base pair enhancer element within the REDFly enhancer d5778 were identified as potential cis-regulatory elements of slp1 and slp2 genes. The distance between the start of GMR35H02 and the end of d5778 is around 27.394 kbp. Other fragments that were screened /cloned are shown as black bars with names on top. (B-B’’’) A 220 bp enhancer element located within the REDfly enhancer u8772 activates GFP expression in medulla neuroblasts in the same pattern as endogenous Slp1 and Slp2 in reporter assays. (C-C’’’) Reporter GFP expression driven by an 850 bp enhancer segment located within the REDfly enhancer d5778 also closely coincides with the expressions of endogenous Slp1 and Slp2, although it is initiated at a slightly later temporal stage than the 220 bp enhancer. (D-D’’’’) A CRISPR-Cas9 edited chromosome 2 with both enhancers deleted (indicated as SlpDED) when placed over the SlpS37A chromosome results in loss of Slp1 and Slp2 expression in medulla neuroblasts in affected flies (n=11). The effect is confined to neuroblasts, as Slp2 expression is retained in both lamina (la) and in surface glial cells (arrows), and Slp1 is also seen in laminar cells. This also confirms that coding sequences of slp1 and slp2 genes are unaffected by the CRISPR-Cas9 editing procedure. Control experiments where SlpDED was placed against a wild-type chromosome 2 (indicated by ‘+’) (E-E’’’’) (n=9) and where the SlpS37A chromosome was placed against a wild-type chromosome 2 (F-F’’’’) (n=11) show normal expression of Slp1 and Slp2 in medulla neuroblasts. Scale bars 20 µm.