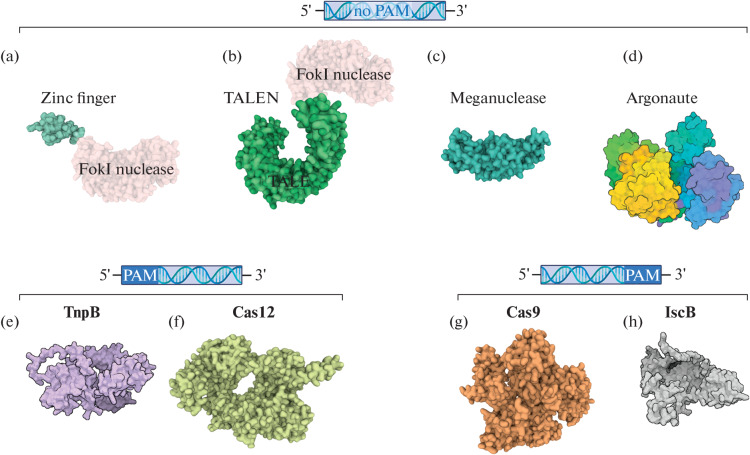
Fig. 1.
Variety of programmable nucleases. (a–d) Endonucleases that do not require a PAM sequence in the target for catalytic activity; (e, f) endonucleases that require the presence of PAM (protospacer adjacent motif) in the 5'-region from the recognition site; (g, h) endonucleases that require the presence of PAM in the 3'-region from the recognition site. (a) Chimeric endonuclease based on the zinc finger DNA-binding domain (PDB ID: 7ZNF) and FokI (PDB ID: 2FOK) (protein structures are combined for illustration); (b) chimeric endonuclease based on the TALE DNA-binding domain (PDB ID: 4HPZ) and FokI (PDB ID: 2FOK) (protein structures are combined for illustration); (c) meganuclease I-AabMI (PDB ID: 4YIT); (d) Argonaute protein can be an RNA- or DNA-guided endonuclease and cleave RNA or DNA (TtAgo, PDB ID: 4NCB); (e) RNA-dependent DNA-endonuclease TnpB (TnpB ISDra2 structure was predicted using the AlphaFold2 algorithm [1]); (f) RNA-dependent DNA-endonuclease Cas12 (PDB ID: 7EU9) [2]; (g) RNA-dependent DNA-endonuclease Cas9 (PDB ID: 6M0X) [3]; and (h) RNA-dependent DNA-endonuclease IscB (the structure was predicted using the AlphaFold2 algorithm [1]).