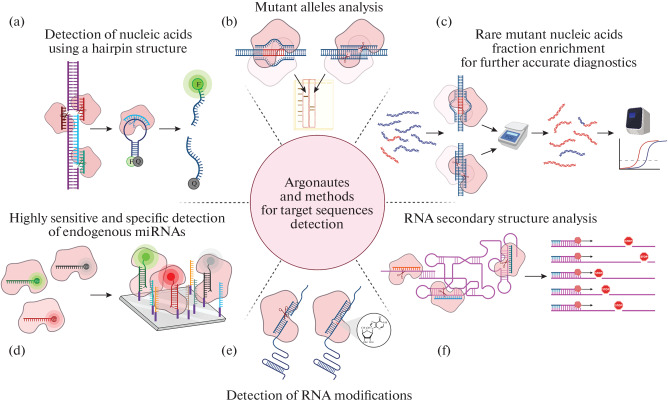
Fig. 4.
Detection of target sequences in biological samples using Argonaute proteins. (a) Detection of nucleic acids using Argonautes. The Argonaute is programmed to cut out the target sequence from the DNA; in addition, an oligonucleotide probe complementary to this sequence is added to the sample; the probe has a hairpin structure, to the ends of which a fluorescent label (F) and a quencher (Q) are attached. During the first round of catalysis, the Argonaute cuts out the target sequence in the sample, after which the Argonaute binds this sequence as a guide and performs the second round of catalysis, as a result of which the fluorescent probe is cleaved. The presence of the target sequence is assessed by increasing the fluorescence intensity [180, 181]. (b) Mutant alleles analysis in biological samples. To detect the mutant allele, DNA is first amplified, and then PCR products are incubated at 98°C with Argonaute loaded with two DNA guides corresponding to one of the variants of the target sequence; then the products are separated in an agarose gel [182]. (c) Detection of rare DNA variants. The mutant DNA sample is incubated with Argonaute loaded with guides corresponding to the wild-type sequence; only mutant DNA remains intact after catalysis; the amount of such DNA can be estimated using Real-time PCR or other methods [183]. (d) For miRNA detection, catalytically inactive Argonaute is loaded with a guide corresponding to the target sequence and containing a fluorescent label; after incubation with miRNAs immobilized on a chip, the fluorescence signal of guides immobilized in a complex with Argonautes and the target miRNA is detected [184]. (e) Detection of RNA modifications [131]. (f) RNA secondary structure analysis. Structured RNAs are incubated with Argonautes loaded with guides to different sequence sites; cleavage sites can be detected directly or by reverse transcription [126, 127, 153].