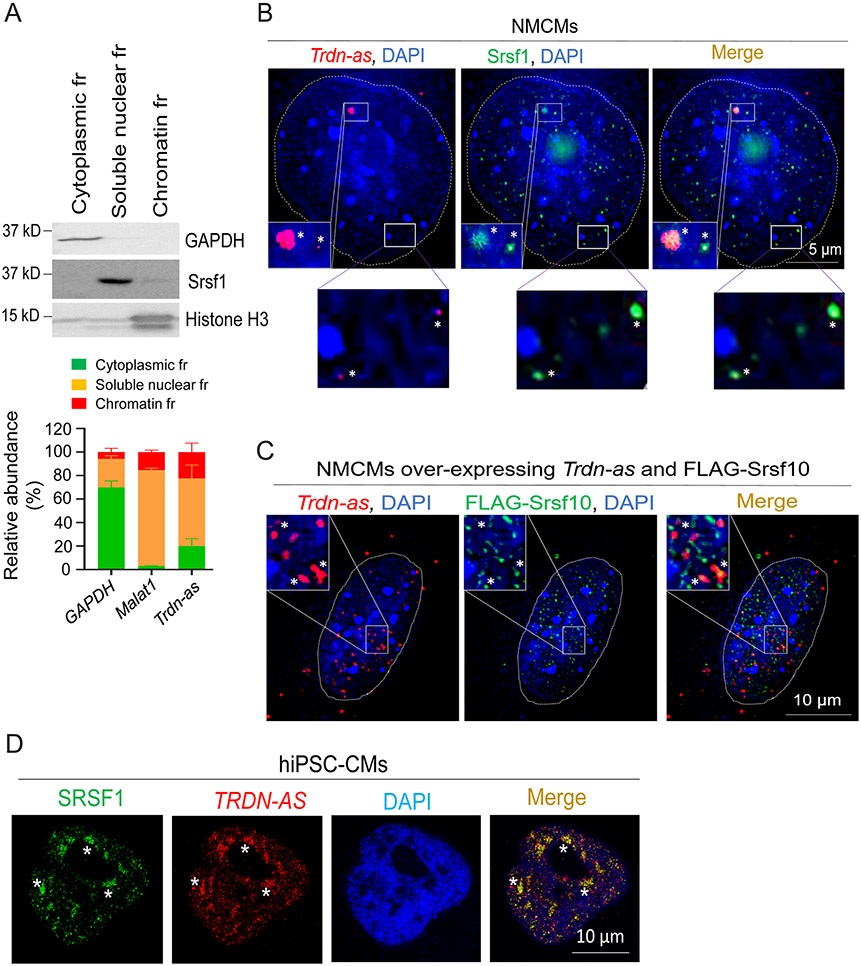
Figure 6. Trdn-as and TRDN-AS colocalize with SR splicing factors in CMs.
A) qPCR analysis of the relative abundance of transcripts from subcellular fractions (fr) of AMCMs. Immunoblotting for GAPDH as a marker for cytoplasm, Srsf1 for soluble nuclear, and histone H3 for chromatin fractions are shown in the top panel. GAPDH and Malat1 RNAs are used as markers for cytoplasmic and soluble nuclear fractions, respectively. N = 3 independent experiments. Data are presented as mean + SD. B-C) Confocal imaging analysis of Trdn-as and Srsf1 in B and over-expressed Trdn-as and FLAG-Srsf10 in C in NMCMs. Endogenous (in B) or over-expressed Trdn-as (in C) were detected by smFISH. In C, NMCMs were infected with adenovirus vectors carrying Trdn-as or FLAG-Srsf10 with a MOI of 5. 48 hours later, over-expressed Trdn-as and FLAG-Srsf10 were analyzed using smFISH and immunostaining, respectively. Nuclei are surrounded by white lines. Insets are enlarged on the right corner. Colocalizations are indicated by stars. D) Confocal imaging analysis of TRDN-AS and SRSF1 in a hiPSC-CM by FISH. TRDN-AS was over-expressed in hiPSC-CMs by adenovirus with a MOI of 5. 48 hours later, TRDN-AS and SRSF1 were analyzed using FISH and immunostaining, respectively. Colocalizations are indicated by stars.