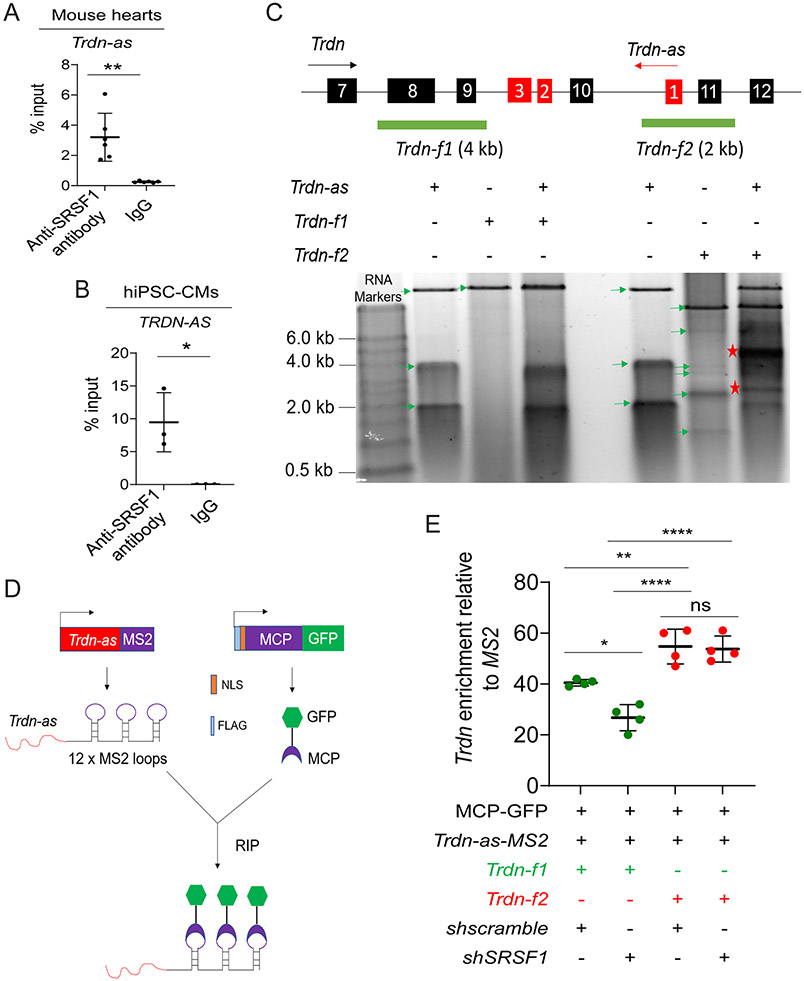
Figure 7. Trdn-as interact with SR splicing factors and Trdn pre-mRNA.
A) RIP-qPCR analysis of the interaction of Trdn-as with Srsf1 in adult mouse hearts. n=6 independent experiments. Data are presented as mean ± SD. Student’s t-test, **p<0.01. B) RIP-qPCR analysis of the interaction of TRDN-AS with SRSF1 in hiPSC-CMs. n=3 independent experiments. Data are presented as mean ± SD. Student’s t-test, *p<0.05. C) Analysis of interactions between Trdn-as and Trdn pre-mRNA by native agarose gel electrophoresis. Trdn and Trdn-as genes schematics are shown on top. Exons of Trdn and Trdn-as are numbered in black and red boxes, respectively. The black and red arrows indicate the direction of Trdn or Trdn-as transcription, respectively. Full-length Trdn-as and two partial fragments of Trdn pre-mRNA, Trdn-f1 and Trdn-f2 were in vitro transcribed. RNA was purified after treatment with proteinase K and RNase-free DNase I. The mixture of Trdn-as and either Trdn-f1 or Trdn-f2 was resolved in native agarose gel electrophoresis. Green arrows indicate individual RNAs. Asterisks indicate RNA-RNA duplex. D) A schematic diagram shows pulldown of lncRNA-associated complexes using the MS2-MCP system. MS2 RNA stem loops strongly interact with the MS2 coat protein (MCP). Trdn-as was fused to 12 x MS2 stem loops (Trdn-as-MS2). Nuclear-localized, FLAG-tagged MCP was fused to GFP (FLAG-NLS-MCP-GFP). Trdn-as associated complexes are precipitated using a GFP antibody. E) qPCR analysis of Trdn pre-mRNA levels in complexes precipitated by the GFP antibody. HEK293 cells were transfected with various combinations of expression plasmids encoding MCP-GFP, shscramble, shSRSF1, MS2, or Trdn-as-MS2. 48 hours later, nuclear extracts of HEK293 cells were incubated with in vitro transcribed Trdn pre-mRNA Trdn-f1 or Trdn-f2. MS2 or Trdn-as-MS2 associated complexes were precipitated by anti-GFP antibody. Trdn pre-mRNA was analyzed using qPCR to amplify Trdn exon 8 for Trdn-f1 or Trdn exon 11 for Trdn-f2, respectively. N = 4 independent experiments. Data are presented as mean ± SD. One-way ANOVA with Tukey’s post hoc test. *p<0.05, **p<0.01, ****p<0.0001, ns, not significant.