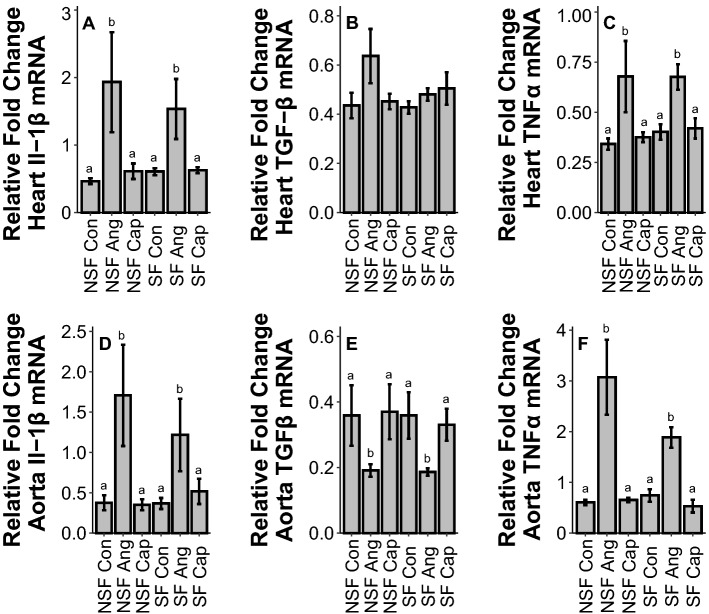
Figure 2.
Effects of sleep fragmentation, RAS manipulation, and their interaction on Il-1β, TGFβ, and TNFα gene expression in circulatory tissues, heart (A, B, C) and aorta (D, E, F). Samples sizes are (A) NSF-Con n = 10, NSF-Ang n = 10, NSF-Cap n = 10, SF-Con n = 9, SF-Ang n = 9, and SF-Cap n = 10, (B) NSF-Con n = 10, NSF-Ang n = 10, NSF-Cap n = 10, SF-Con n = 10, SF-Ang n = 8, and SF-Cap n = 10, (C) NSF-Con n = 10, NSF-Ang n = 9, NSF-Cap n = 10, SF-Con n = 10, SF-Ang n = 9, and SF-Cap n = 9, (D) NSF-Con n = 11, NSF-Ang n = 10, NSF-Cap n = 9, SF-Con n = 9, SF-Ang n = 9, and SF-Cap n = 10, (E) NSF-Con n = 11, NSF-Ang n = 10, NSF-Cap n = 10, SF-Con n = 9, SF-Ang n = 8, and SF-Cap n = 10, (F) NSF-Con n = 11, NSF-Ang n = 10, NSF-Cap n = 9, SF-Con n = 9, SF-Ang n = 9, and SF-Cap n = 10. All data were analyzed using a two-way ANOVA and Tukey’s HSD post hoc tests. Data shown as means ± 1 SE for each group and differing lowercase letters denotes p < 0.05.