Figure 1.
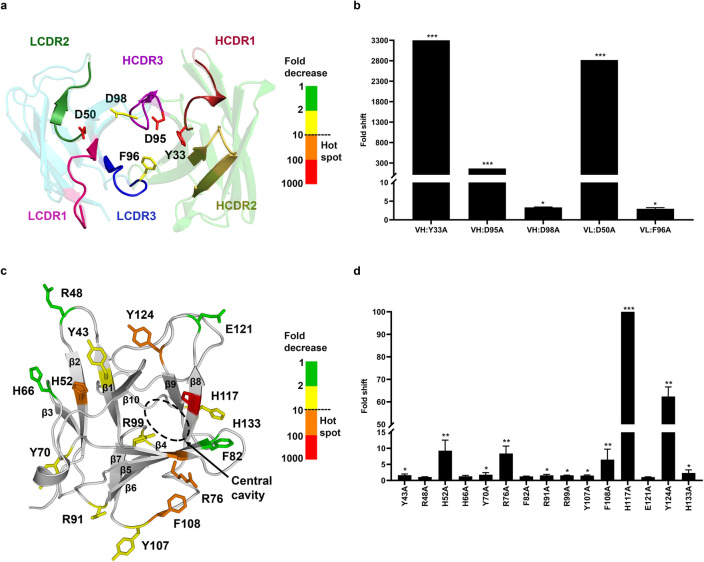
Paratope and epitope scanning for hotspot identification. Fifty-six Burosumab residues that were solvent exposed and covered all CDR loops were mutated to alanine and tested individually for binding to wild-type FGF23 using spot ELISA (Supplementary Tables S2 and S3). For each alanine mutant, the fold decrease in binding was calculated by comparing A450 of each mutant with wild-type Burosumab. The five key affinity altering residue positions (VH:Y33, VH:D95, VH:D98, VL:D50, VL:F96) were identified and mapped onto FGF23-binding interface (a). The residue coloring indicates the fold shift in binding relative to WT Burosumab: green: 1–2; yellow: 2–10; orange: 10–100; red: 100–1000. These five positions were considered as constraints for computational docking. (b) Fold decrease in binding for a representative set of mutations was shown. Degree of fold decrease was denoted by asterisk (*) as compared to the wild type: 2 to tenfold (*), > 10 to 100 fold (**), and > 100 fold (***). The homology model of Burosumab was built using Rosetta Antibody 3 provided by ROSIE server. CDR numbering was based on Chothia’s definition. For epitope scanning, fifteen solvent-exposed FGF23 residues that covered every domain of N-terminal domain of FGF23 were mutated to alanine and tested for binding to wild-type Burosumab. Fold decrease in binding was calculated from indirect ELISA of FGF23-Burosumab interaction by comparing a Kd value (nM) of each mutant with wild-type FGF23. Five epitope residues: H52, R76, F108, H117, Y124 with nearly a 10-fold decrease in binding or more upon alanine mutation were mapped onto FGF23 structures (c,d). These key epitope residues appeared to be in close proximity to the D2 and D3 domains of FGF23 receptor (FGFR) (Supplementary Fig. S1). N-terminal domain of FGF23 structure was sourced from PDB ID: 5W21. Structures were illustrated by PyMOL version 2.2.0. The graphs were created by GraphPad Prism version 8.4.3.