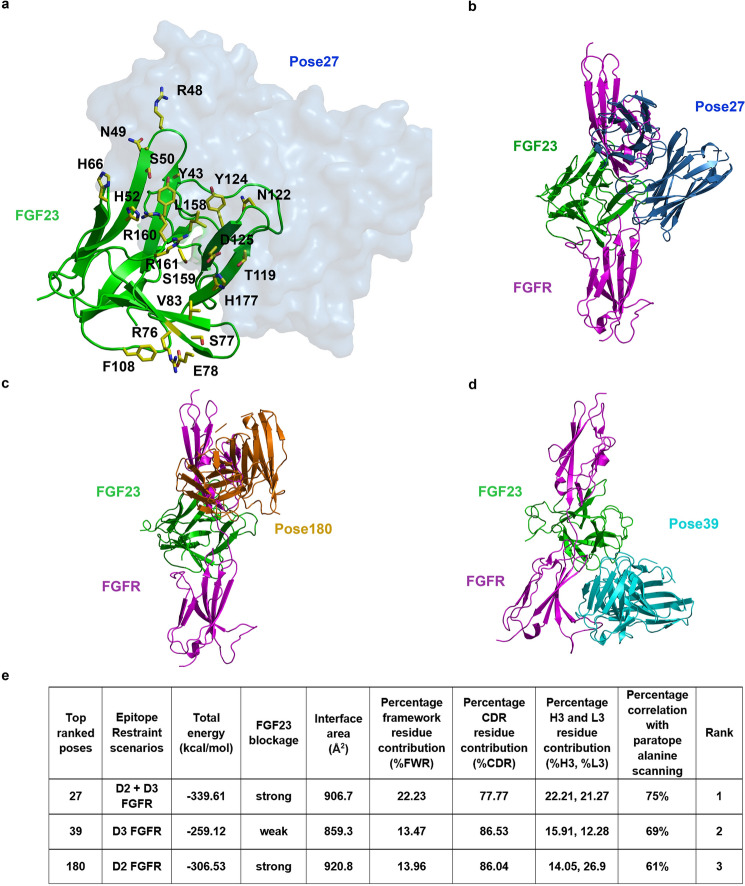
Figure 2.
Computational docking and model assessment. HADDOCK2.4 was run using default parameters with the homology model of Burosumab Fv (RosettaAntibody3) and the unbound crystal structure of FGF23 (PDB ID: 5W21). The identified paratope and epitope residues from alanine scanning were used as active residue restraints during the docking process. Pose27 was seen to interact with a number of FGFR-interacting FGF23 residues (yellow sticks), highlighting the FGF23:FGFR blocking potential of the antibody (a). The top pose from each docking scenario was superimposed with FGF23-FGFR complex (PDB ID: 5W21) using PyMOL version 2.20 (b–d). Table in panel (e) shows the assessment of the top three poses using the seven structural criteria. Amongst the three poses, pose27 had the most favorable structural characteristics: (1) high FGF23 blockage, (2) the lowest binding energy, (3) the best correlation with paratope alanine scanning data, (4) acceptable interface area (> 800 Å2), (5) reasonable %FWR for lock-and-key binding given a comparable size between FGF23 and Burosumab Fv, (6) acceptable percent contribution of CDR residues at the interface (%CDR), and (7) highest contribution of H3 and L3.