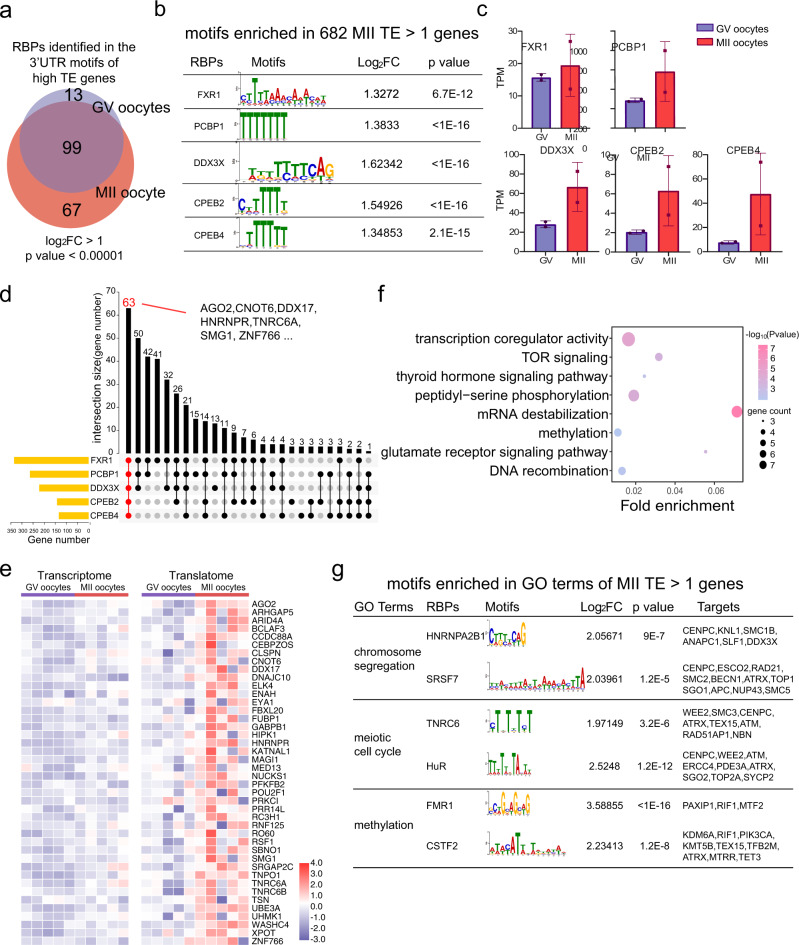
Fig. 3. High TE genes carry RNA-binding-protein motifs at their 3’UTRs with high frequency.
a Venn plot of putative RBPs identified in 3’UTR motifs of high TE genes in GV and MII oocytes, log2Fold change > 1, p < 0.00001, one-sided t-test (see the “Methods” section). Blue circle covers RBPs bound to 3’UTR of high TE in GV oocytes. Red circle covers RBPs bound to 3’UTR of high TE in MII oocytes. Intercepted area indicates RBPs bound to 3’UTR of high TE in both GV and MII. b 5 RBPs with the high significant values and their binding motifs enriched in MII TE > 1 gene (p-value, one-sided t-test). c The translational expression levels in GV and MII of the top 5 RBPs shown in b. Data from two biological replicates of 10 human oocytes. Error bar denotes standard deviation (SD). d Upset plot showing the number of the overlapped targeted genes containing the five RBP motifs. 63 genes including AGO2, CNOT6 are potentially co-regulated by the 5 RBPs. e T&T heatmap of representative genes potentially co-regulated by 5 RBPs. f Representative GO terms of the genes co-regulated by the 5 RBPs (p-value, hypergeometric test). g motifs enriched in GO terms of MII TE > 1 genes (p-value, one-sided t-test).