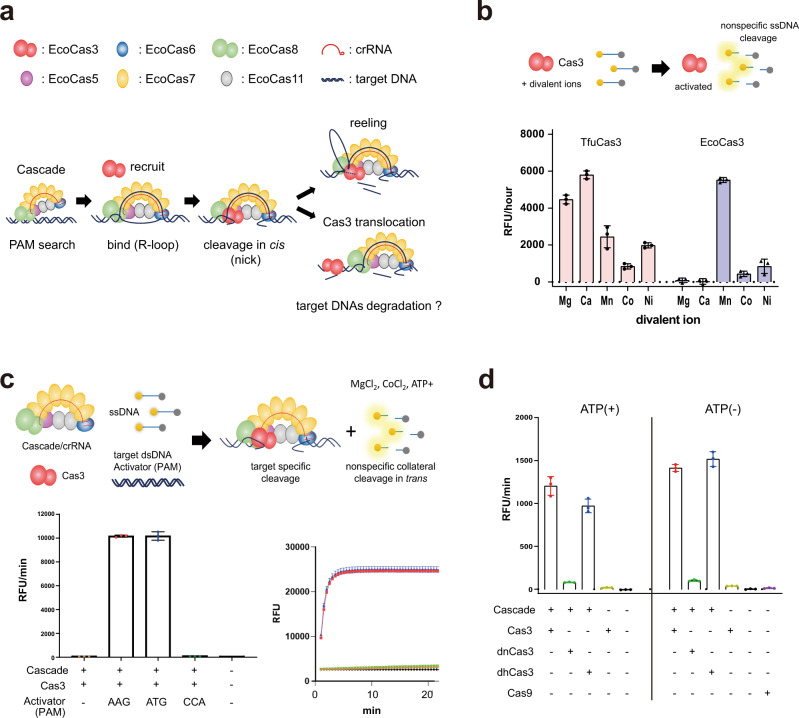
Fig. 1. In vitro reconstituted EcoCas3-EcoCascade/crRNA complex cleaves nonspecific ssDNA in trans.
a Schematic depiction of the known type I CRISPR interference mechanism. b, c Electrophoretic mobility shift assay (EMSA) and DNA degradation assay. b Activation of EcoCas3 and TfuCas3 by divalent metal ions (Mg2+, Ca2+, Mn2+, Co2+, and Ni2+). Fluorescent dye-quencher (FQ)-labeled ssDNA probes measured promiscuous ssDNA cleavage activity. RFU: relative fluorescence unit. c Collateral ssDNA cleavage activity measured by incubation of EcoCas3-EcoCascade/crRNA complex with a 60 bp dsDNA Activator containing a target sequence flanked by a PAM and an FQ-labeled ssDNA probe in reaction buffer containing MgCl2, CoCl2, and ATP for 10 min at 37 °C. Quantitatively represented by RFU per min (left) or RFU at 10 min (right). d EcoCas3 HD domain H74A (dead nuclease mutant, dnCas3), abolished collateral cleavage activity, while SF2 motif III S483A/T485A (dead helicase mutant, dhCas3) showed collateral cleavage activity. Collateral activity in ATP reaction buffer (+) was at the same level as that in ATP-free buffer (−) for wild-type EcoCas3 and the dhCas3 mutant. SpCas9 did not exhibit any collateral cleavage activity. Data in b–d are presented for n = 3 independent measurements and mean value, error bars represent SD values. Source data are provided as a Source Data file.