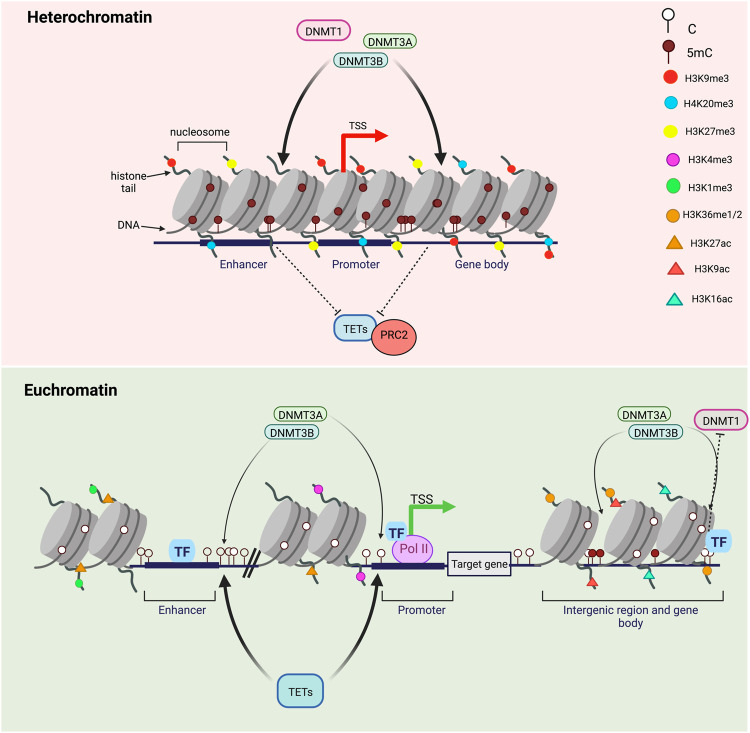
FIGURE 3.
DNA methylation turnover in heterochromatin versus euchromatin. Transcriptionally silent genes are found in highly-condensed chromatin regions called heterochromatin and marked by repressive histone marks H3K9me3, H3K27me3 and H4K20me3. DNA methylation levels in heterochromatin are high and DNA methylation turnover rates are usually low. CpGs in heterochromatic regions are highly methylated by DNMTs and inaccessible by TET proteins. On the opposite, transcriptionally active genes are associated with a less-condensed, nucleosome-depleted and accessible chromatin known as euchromatin. In transcriptionally active regions, gene promoters are enriched in H3K4me3 and H3K27ac, enhancers in H3K4me1 and H3K27ac and gene bodies in H3K9Ac and H3K36me. Highly methylated CpGs within highly transcribed genes, gene bodies and nearby regulatory regions (enhancers and promoters) are subjected to high methylation turnover rates while hypermethylated CpGs within intergenic regions have lower TET and DNMT engagement. Transcription factors binding can regulate both methylation by DNMTs and demethylation by TETs.