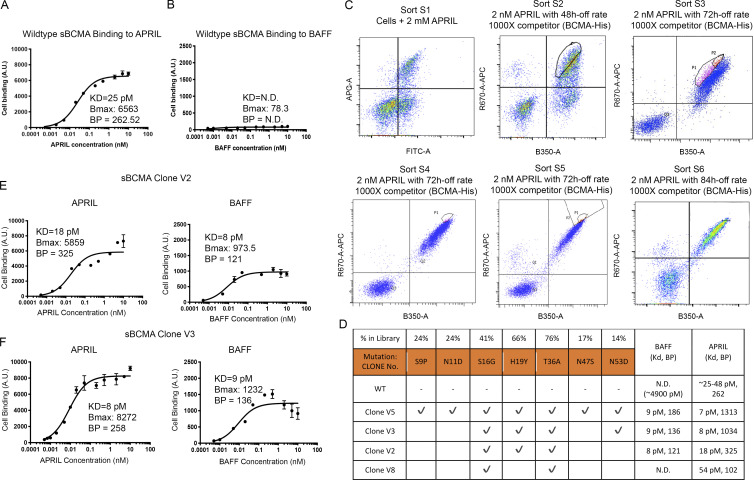
Figure 4.
Engineering high-affinity decoy receptor fusion protein against APRIL and BAFF. (A) Flow cytometry–based binding curve showing yeast-displayed wild-type sBCMA binding to increasing concentrations of APRIL. Calculated KD, Bmax, and binding potential (BP) is also shown. A.U., arbitrary units. (B) Flow cytometry–based binding curve showing yeast-displayed wild-type sBCMA binding to increasing concentrations of BAFF. Calculated KD, Bmax, and BP is shown. (C) Overlaid flow cytometry dot plots representing sorting strategies of yeast-displayed sBCMA library binding to 2 nM APRIL in six consecutive sorts with 48–84 h off-rate binding and 1,000× competitor. Gated populations are collected from each sort and propagated for the next sorting round. (D) Binding affinities to APRIL and BAFF of wild-type sBCMA and selected mutant sBCMA clones. Conserved amino acid mutations were identified in mutant clones, and the frequency of occurring mutations is also listed. (E) Binding curve of high-affinity mutant sBCMA clone V2 binding to increasing concentrations of APRIL (left) and BAFF (right). Calculated KD, Bmax, and BP is shown. (F) Binding curve of high-affinity mutant sBCMA clone V3 binding to increasing concentration of APRIL (left) and BAFF (right). Calculated KD, Bmax, and BP is shown.