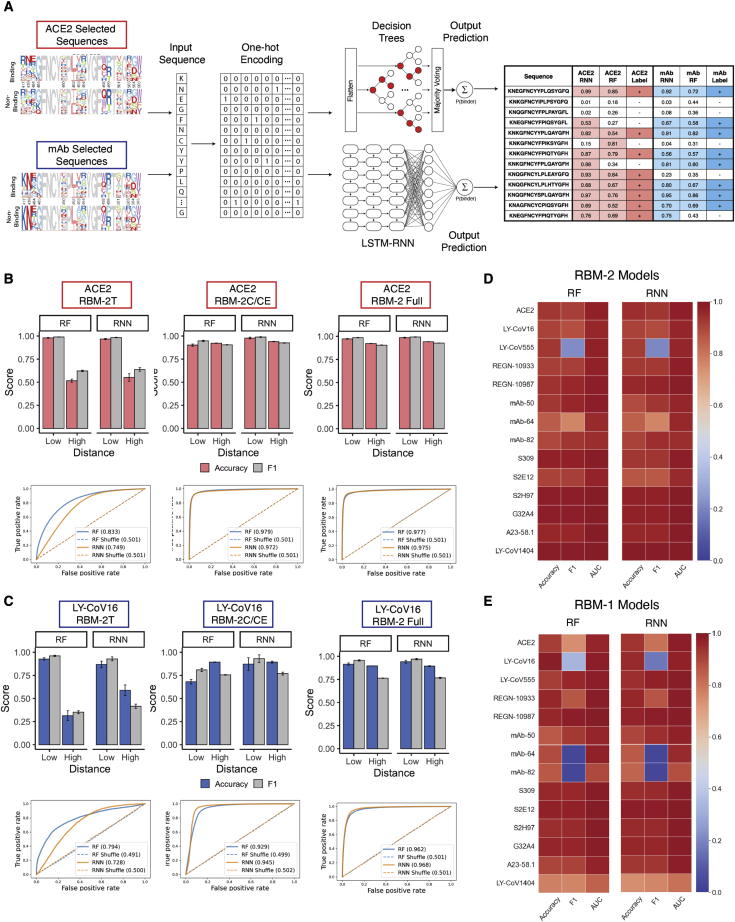
Figure 3.
Training and testing of machine and deep learning models for prediction of ACE2 binding and antibody escape based on RBD sequence
(A) Deep sequencing data from ACE2 and monoclonal antibody (mAb) selections is encoded by one-hot encoding and used to train supervised machine learning (e.g., random forest [RF]) and deep learning models (e.g., recurrent neural network [RNN]). Models perform classification by predicting a probability (P) of ACE2 binding or non-binding and mAb binding or escape (non-binding) based on the RBD sequence.
(B and C) Performance of RF and RNN models trained on 2T, 2C, or Full ACE2 or LY-CoV16 binding data shown by accuracy, F1, and receiver operating characteristic (ROC) curves. Models are evaluated by rounds of external cross-validation (n = 5), with mean performance displayed and standard deviation indicated by error bars. Low and high distance sequences are defined as those ≤ED5 and ≥ED6 from Wu-Hu-1 RBD, respectively.
(D and E) Accuracy, F1, and AUC of all 13 mAb models trained on RBM-2 and RBM-1 data, evaluated on both low and high distance test sequences.
See also Figures S3 and S4 and Table S4. Detailed sequences used as the training data for individual models, Table S6. Machine and deep learning model predictions compared to susceptibility data from the Stanford Database.