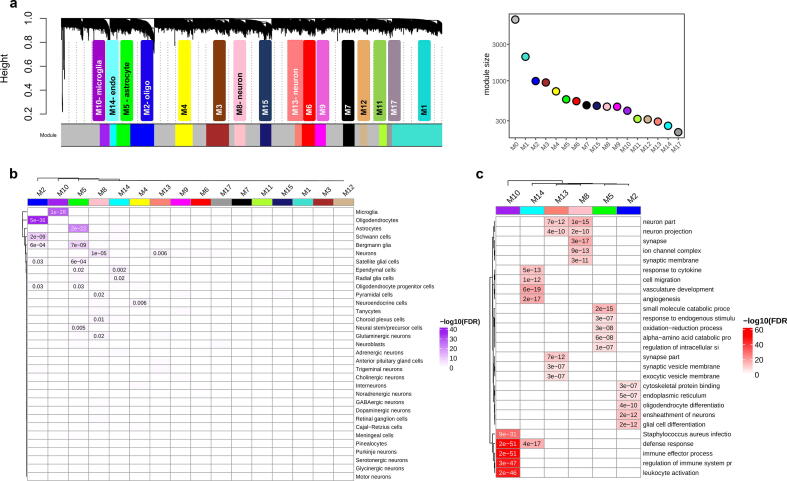
Fig. 4.
Cross-condition co-expression modules identified by network analysis. (a) A dendrogram plot displaying co-expression modules obtained from the topological overlap of 15,819 shared genes between conditions. Each color represents an individual module and the grey color (M0) contains genes that are not included in a specific module. The corresponding plot on the right side shows the number of genes within each module. (b) Enrichment of co-expression modules for brain cell types, measured by comparing genes within each module to the brain single-cell dataset from PanglaoDB [56] (see also Supplementary Fig. S21a). (c) Heatmap plot of gene ontology enrichment for cell-type-specific modules using top five significant pathways for each module. The color key shows -log10(FDR).