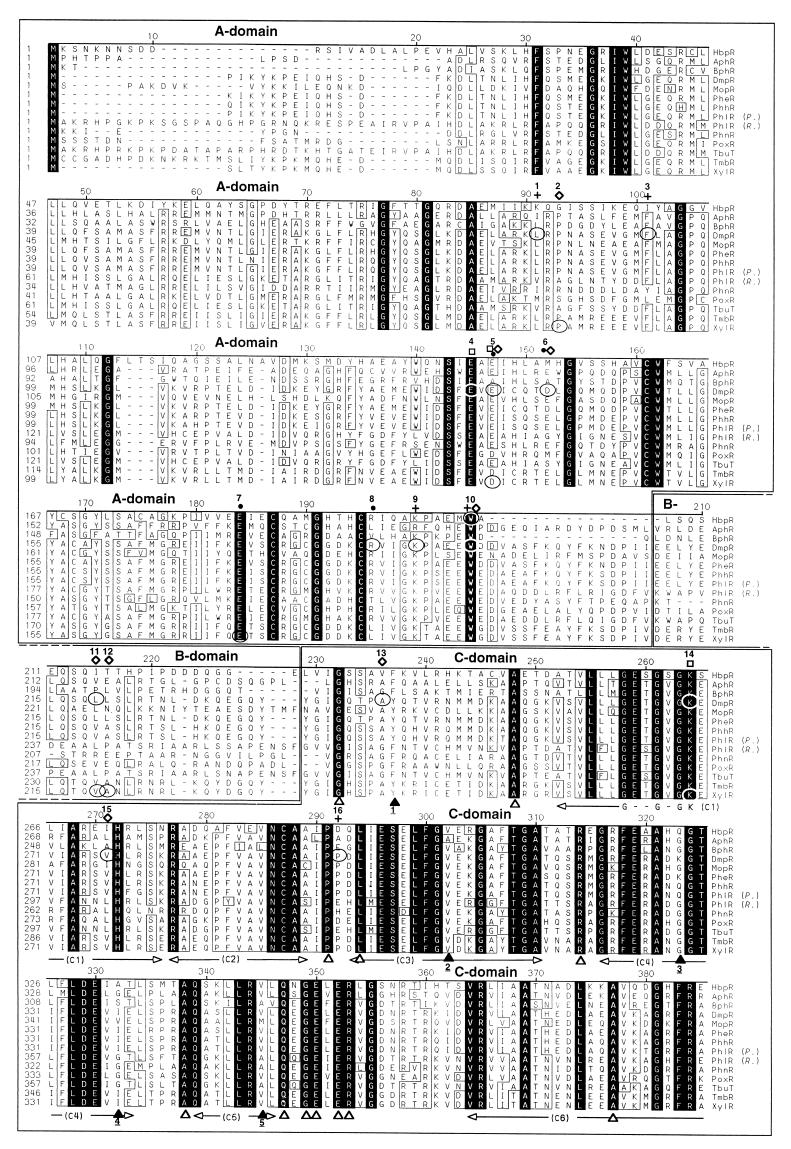
FIG. 4.
Alignment of the predicted amino acid sequences of the HbpR protein and the 13 most similar members of the NtrC family of transcriptional activators in the nonredundant (NR) protein sequence database. The alignment was made with the program MegAlign from the Lasergene package (DNASTAR, Inc.) and further improved manually. The ruler shown above the sequences corresponds to the amino acid sequence of the HbpR protein. Conserved amino acid residues in all proteins are indicated against a black background, whereas residues conserved in at least 10 of 14 proteins appear boxed. Gaps introduced to maximize the alignment are dashed. The regions corresponding to the different domains are boxed, and their names are displayed above the sequences. The borders of the A, C, and D domains in the different proteins were set according to those proposed for the XylR protein (25). The sections C1 to C7, shown below the sequences, correspond to the conserved regions within the C domain as described by Morett and Segovia (40). The Walker A motif within regions C1 (55, 69), involved in ATP binding, and a putative helix-turn-helix in the D domain (40) are indicated below the sequences. U represents hydrophobic residues. White triangles below the sequences mark fully conserved residues within the C and D domains of the XylR/DmpR subclass members but not fully conserved in an alignment made by Morett and Segovia, which included 27 members of the NtrC family, including XylR (40). Black triangles below the sequence mark positions which were not conserved in at least 10 of 14 proteins but appeared to be conserved in the alignment made by Morett and Segovia (40). Underlined numbers below the black triangles indicate which residues replaced the conserved ones: 1, F or Y instead of L or M; 2, A or V instead of H; 3, G, K or Q instead of only G: 4, A, G, I, S, or V instead of only G; 5, A, M, or V instead of I or V; 6, A or P instead of only P; 7, C, L, or M instead of L or M; 8, L, M or V instead of L or M. Encircled residues mark the positions of described point mutations within XylR/DmpR subclass members. The phenotypes of described point mutations is indicated above the sequence: black dots, altered effector specificity (ALT); open diamonds, constitutive or semiconstitutive transcription-promoting activity (CON); empty squares, abolition of transcription-promoting activity (NON); crosses, suppressor mutation of a semiconstitutive mutation (SUP); or a combination of these symbols. Numbers above these symbols are used to describe all known mutations for the corresponding position and their resulting phenotype(s): 1, DmpR-L83P, SUP selected for DmpR-E135A (42); 2, XylR-P85S, CON (13); 3, DmpR-F93L, SUP selected for DmpR-E135D (42); 4, DmpR-E133K, NON (61); 5, DmpR-E135A, ALT CON (61); DmpR-E135D, ALT CON (61); DmpR-E135K, ALT (46); DmpR-E135R, ALT (61); XylR-D135E, NON (53); XylR-D135N, CON (13); XylR-D135Q, CON (53); 6, DmpR-D140K, ALT CON (61); 7, XylR-E172K, ALT (12); 8, DmpR-R184W, ALT (12); 9, DmpR-K188E, SUP selected for DmpR-E135D (42); 10, HbpR-W205R, CON (this study); DmpR-W193R, SUP for DmpR-V276A (42); 11, DmpR-L219P, CON (61); XylR-V219DA220P (double mutation), CON (18); 12, XylR-V219DA220P (double mutation), CON (18); 13, DmpR-A241T, CON (61); 14, DmpR-G268S, NON (42); XylR-G268N, NON (48); 15, DmpR-V276A, CON (61); DmpR-V276G, CON (61); 16, DmpR-P297R, SUP selected for DmpR-E135D (42); 17, DmpR-F424L, CON (61); 18, XylR-R453H, NON (48). The database entries for the different proteins are given in the legend of Fig. 6.