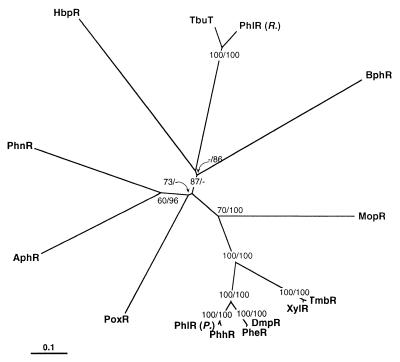
FIG. 6.
Single unrooted phylogenetic tree of the HbpR amino acid sequence with 13 members of the XylR/DmpR subgroup. The tree was obtained by protein distance calculations (PROTDIST, Kimura setting) and subsequent neighbor-joining analysis (NEIGHBOR; both programs from the software package Phylip version 3.5 [17]) on a subalignment of 616 aa including gaps. Values at the forks indicate the number of times that particular branching occurred among 100 resamplings of the data set (generated with bootstrapping) as determined by consensus neighbor-joining analysis (first value) or maximum-parsimony analysis (second value). Missing values indicate that particular branching to be absent in the consensus tree. The consensus tree was displayed by using TREEVIEW (45). The bar indicates 0.1 change per site. The origins of the different proteins are as follows: AphR from Comamonas testosteroni TA441 (GenBank accession no. AB006480) (4); BphR encoded on plasmid pNL1 from Sphingomonas aromaticivorans F199 (GenBank accession no. AF079317) (52); DmpR encoded on plasmid pVI150 from Pseudomonas sp. strain CF600 (GenBank accession no. X68033) (59); HbpR from P. azelaica HBP1 (GenBank accession no. U73900) (this study); MopR from Acinetobacter calcoaceticus NCIB8250 (GenBank accession no. Z69251) (56); PheR from P. putida BH (GenBank accession no. D63814); PhhR from P. putida P35X NCIB9869 (GenBank accession no. X79599) (43); PhlR encoded on plasmid pPGH1 from P. putida H (GenBank accession no. X91145) (41) in the figure indicated as PhlR (P.); PhlR from a Ralstonia eutropha JMP134 derivative (GenBank accession no. AF065891), in the figure indicated as PhlR (R.); PhnR from Burkholderia sp. strain RP007 (GenBank accession no. AF061751) (35); PoxR from R. eutropha E2 (GenBank accession no. AF026065) (24); TbuT from R. pickettii PKO1 (GenBank accession no. U72645) (10); TmbR from P. putida TMB (GenBank accession no. U41301) (16); XylR encoded on plasmid pWW0 from P. putida mt-2 (GenBank accession no. M20635) (25).