Abstract
In the Northeast of Brazil, Ceará was the second state most impacted by COVID-19 in number of cases and death rate. Despite that, the early dynamics of the pandemic in Ceará was not yet well understood due the low genomic surveillance of SARS-CoV-2 in 2020. In this study, we analyze the circulating lineages and the genomic variation of the virus in Ceará state. Thirty-four genomes were sequenced and combined with sequences available in GISAID database from March 2020 to June 2021 to compose the study dataset. The most prevalent lineages detected were B.1.1.33, in 2020, and P.1, in 2021. Other lineages were found, such as P.2, sublineages of P.1, B.1, B.1.1, B.1.1.28 and B.1.212. Analyzing the mutations, a total of 202 single-nucleotide polymorphisms (SNPs) were identified among the 34 genomes sequenced, of which 127 were missense, 74 synonymous, and one was a nonsense mutation. Among the missense mutations, C14408T, A23403G, T27299C, G28881A G28883C, and T29148C were the most prevalent within the dataset. Although SARS-CoV-2 sequencing data was limited in 2020, our results could provide insights to better understand the genetic diversity of the circulating lineages in Ceará.
Keywords: COVID-19, Genome sequences, Mutations
1. Introduction
In December 2019, the disease known as COVID-19, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), was detected in Wuhan, China (Wang et al., 2020; Zhu et al., 2020), and was later declared as a pandemic by the World Health Organization (WHO) in March 2020. Since then, the virus has spread rapidly resulting in more than six million deaths worldwide as of August 2022 (WHO, 2022). In Brazil, the first confirmed contagion of SARS-CoV-2 was in late February 2020 in the state of São Paulo (de Jesus et al., 2020). After this, the country rapidly became one of the epicenters of the pandemic with lineages B.1.1.28 and B.1.1.33 being the most prevalent in the early epidemic phase (Resende et al., 2021). In late 2020, two variants, Zeta (P.2) (Voloch et al., 2020) and Gamma (P.1) (Faria et al., 2021a), descendants of lineage B.1.1.28, emerged and were associated with the second phase of the pandemic. With a population of over 9.2 million people, Ceará is an economically relevant state in Brazil, with a strong travel industry and a high-traffic airport located in its capital, Fortaleza. Ceará has the second highest number of cases in the northeast of Brazil, with 27,482 deaths registered (http://covid.saude.gov.br, accessed on 12 August, 2022). The first reports of COVID-19 in Ceará dated from March 2020. From the early pandemic, genomic surveillance has been an efficient tool to trace variants of SARS-CoV-2 and to study the virus import and spread. Furthermore, in 2020, before a strong genomic surveillance service was established in Ceará, only few SARS-CoV-2 genomes were sequenced to infer the lineages that were circulating in the first months of pandemic. After the first SARS-CoV-2 genome sequence was made available, in January 2020 (Wu et al., 2020), over 12 million genomes were sequenced and shared on the Global Initiative on Sharing All Influenza Data (GISAID) database (Khare et al., 2021), allowing the identification of the virus lineages worldwide. Mutations have emerged throughout the virus genome, but those related to gene S are more relevant, once its product, the Spike protein, is directly involved in the host cell entrance process (Fung and Liu, 2019). For example, a single amino acid change from aspartic acid to glycine at position 614 of protein Spike (D614G) became dominant in a short time, and was associated with increased transmission of the virus (Korber et al., 2020). Knowledge of new mutations and circulating lineages is essential for decision making on measures to contain the pandemic, since each variant may influence in the pathogenicity and transmissibility of the virus (Lauring and Hodcroft, 2021; Saito et al., 2021; Wang et al., 2021). In this study, we used thirty-four SARS-CoV-2 genome sequences to investigate the circulating lineages and detect mutation patterns to better understand the dispersion and evolution of the SARS-CoV-2 in the early phase of the epidemic in Ceará. Sequences from 2020 were used to determine the circulating lineages in this year, before the strike of the second wave, during which Gamma lineages were most prevalent. In addition, we highlight the importance of monitoring SARS-CoV-2 lineages through genomic surveillance as a measure to contain the pandemic.
2. Material and methods
2.1. Ethical Aspects
This research was approved by the Federal University of Ceará (UFC) Ethics Committee (CEP/CAAE: 31453320.7.0000.5054) and the Brazilian Ministry of Health SISGEN (A29A4F4).
2.2. Sample selection and viral detection by RT-qPCR
From 5,449 samples used to perform SARS-CoV-2 detection for diagnosis from July 2020 to June 2021, 34 clinical RT-qPCR positive samples with the lowest cycle threshold of each month were chosen for this study. The diagnostic procedure was performed at the Laboratory of Pharmacogenetics (FARMAGEN) in the Drug Research and Development Center (NPDM), Federal University of Ceará (UFC). Nasopharyngeal swabs were confirmed as positive for SARS-CoV-2 using iTaq Universal Probes One-Step Kit (Bio-Rad, USA) on a QuantStudio 5 instrument (Thermo Fisher Scientific, USA). The protocol used was established by the Centers for Disease Control and Prevention (CDC, Atlanta, USA). To detect the presence of SARS-CoV-2, N1 and N2 genes from the viral Nucleocapsid were used, and the human RNase P gene was used as an internal control.
2.3. Nucleic acid extraction, library preparation, and sequencing
The viral RNA was extracted from 140 μL of clinical samples using QIAamp Viral RNA Mini kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. The libraries were prepared using AmpliSeq Plus or COVIDSeq kits (Illumina, San Diego, USA), according to the manufacturers’ protocols. AmpliSeq Plus libraries were purified with AMPure XP magnetic beads (Beckman Coulter, Brea, USA). Libraries were quantified using High Sensitivity dsDNA quantification kit with Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, USA), and the mean fragment size was analyzed by TapeStation 4150 with DNA HS D1000 kit (Agilent, Santa Clara, USA). Library concentration was calculated and diluted to 4 nM. Libraries were pooled, denatured, and diluted to a final concentration of up to 12 pM and sequenced on a MiSeq platform with MiSeq Reagent kit v2 (300-cycle) (Illumina, San Diego, USA) to generate reads of 2 × 150 bp. The DNA sequencing was performed at the Genomics and Bioinformatics Center (CeGenBio), from NPDM. The sequences are available in the GISAID database (https://gisaid.org/) (Supplementary Table S1).
2.4. Data analysis
Sequencing data were inspected for overall quality with FastQC v0.11.9 (Andrews, 2010) and trimmed with Trimmomatic v0.38 (Bolger et al., 2014). Good quality reads were then processed and analyzed to variant calling and lineage classification. Reads were aligned to SARS-CoV-2 reference genome (NCBI acc. ID NC_045512.2) using BWA v0.7.17-r1188 (Li and Durbin, 2009). SortSam v2.18.29 from Picard Tools (http://broadinstitute.github.io/picard/) was used to sort the alignment, and samtools v1.11 (Danecek et al., 2021) was used for indexing. GATK v.4.1.9.0 (Van der Auwera and O'Connor, 2020) called, selected and filtered variants according to the alignment of samples’ reads to the reference genome. Variants were annotated according to the coding sequences of the reference using snpEff v5.0e (Cingolani et al., 2012a) and the resulting VCF file was converted into a table using SnpSift v.4.3t (Cingolani et al., 2012b). In order to classify the samples into pango lineages we assembled samples’ reads with Skesa v.2.1 (Souvorov et al., 2018) generating contigs for each genome sample. We used RagTag v.2.0.0 (Alonge et al., 2019) to order and orient contigs generated by Skesa. Pangolin v.3.1.11 (github.com/cov-lineages/pangolin) was used to assign the pango lineage to each genome. Complementary analysis was performed with DRAGEN COVID Lineage app v.3.5.8 from Illumina`s BaseSpace (basespace.illumina.com). Additional 1,619 SARS-CoV-2 genome sequences collected from March 2020 to June 2021 in the state of Ceará were downloaded from GISAID (gisaid.org/EPI_SET_20220721sn) to analyze the prevalent lineages circulating in this period (Supplementary Table S2).
2.5. Maximum Likelihood phylogenetics
A Maximum Likelihood (ML) phylogenetic tree was generated with the 34 genome sequences and representants of all Variants of Interest (VOI) and Variants of Concern (VOC) with collection dates within the study period – from July 2020 to June 2021. The final dataset was composed of 121 SARS-CoV-2 genome sequences, and included the Wuhan reference sequence (NCBI acc. num. NC_045512.2) and 86 genome sequences downloaded from GISAID, with collection sites in all continents, in Brazil and in Ceará (Supplementary Method). Multiple alignment was performed with MAFFT v7.505 (Katoh and Standley, 2013), and the ML tree construction was made with IQ-TREE multicore version 2.2.0.5 (Minh et al., 2020), using the GTR+I+F+R3 substitution model and SHaLRT with 1,000 replicates as indicated by ModelFinder (Kalyaanamoorthy et al., 2017). FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) was used to create the visualization.
3. Results
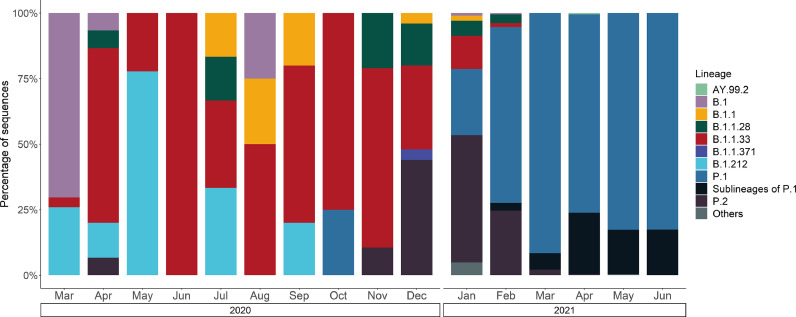
We sequenced 34 SARS-CoV-2 samples from Ceará, Brazil, which were later classified into pango lineages (Supplementary Table S1). Among these, 25 were collected from July to December 2020 and nine from January to June 2021. The genomic sequences were classified into seven different lineages, being B.1.1.33 (n=15, 44.12%) the most frequent, followed by P.1 (n=8; 23.53%), B.1.212 (n=3, 8.82%) and B.1.1 (n=3, 8.82%), B.1.1.28 (n=2, 5.88%), P.2 (n=2, 5.88%), and B.1 (n=1, 2.94%). Moreover, 1,619 sequences from GISAID from the state of Ceará were added to the dataset to determine the circulating SARS-CoV-2 lineages from March 2020 to June 2021, resulting in a dataset of 1,653 sequences. Considering this dataset, the most prevalent lineages were P.1 (72.41%), P.2 (7.38%), P.1.10 (6.05%) and B.1.1.33 (3.69%). Other lineages were found, although in less proportion (Fig. 1 ).
Fig. 1.
Prevalence of SARS-CoV-2 genome sequences for each month from Ceará.
Lineages B.1 (1.39%) and B.1.212 (1.15%) were not constantly represented throughout the months, being sampled mainly up to September 2020. Lineage B.1.1.33 was observed in all months from 2020 and in early 2021, but was not observed from March 2021 on. Even though the lineage B.1.1.28 (1.45%) was dominant in the early pandemic phase in Brazil, it did not have a high prevalence in Ceará in any sampled month, according to the data available. P.2 lineage (VOC Zeta) was first observed in April, although was not again sampled until November, and presented higher prevalence from December 2020 to January 2021, reaching 46.23% in January. Lineages P.1, P.1.10 and other sublineages of P.1 became more prevalent starting from February 2021 and remained frequent until the end of the study period. Notably, we have found one genome sequence belonging to Gamma (P.1) variant in October 2020.
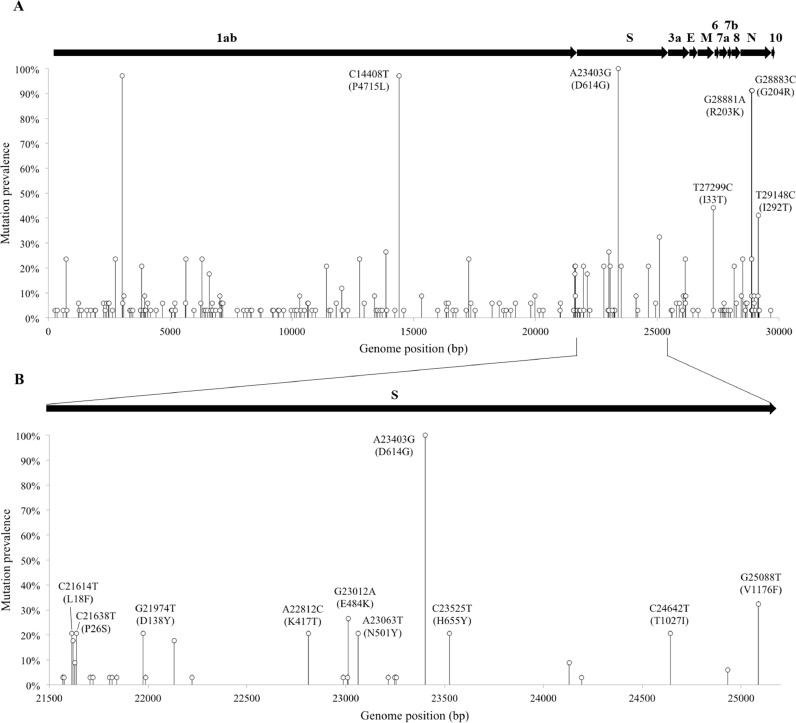
The occurrences of mutations in the 34 SARS-CoV-2 genomes sequenced were also assessed. A total of 202 single-nucleotide polymorphisms (SNPs) were found, among which 127 were missense (non-synonymous), 74 synonymous and one nonsense (Supplementary Table S3). Missense mutations with over 40% of prevalence within the dataset were C14408T (P4715L) in ORF1ab; A23403G (D614G) in gene S; T27299C (I33T) in ORF6; and G28881A (R203K), G28883C (G204R) and T29148C (I292T) in gene N (Fig. 2 a). All high-prevalence mutations found in the dataset are signature to at least one lineage (Supplementary Table S4), with the exception of ORF1ab:P4715L. In gene S, 23 missense SNPs were identified and, among which, the mutation D614G was detected in 100% of the sequenced genomes (Fig. 2b). Missense SNPs in the gene S with a frequency of at least 20% among the genomes analyzed also included C21614T (L18F), C21638T (P26S), G21974T (D138Y), A22812C (K417T), G23012A (E484K), A23063T (N501Y), C23525T (H655Y), C24642T (T1027I), and G25088T (V1176F). The only deletion found within the dataset was the VOC Gamma synapomorphic deletion in position 11,287 (S3675-F3677_), which is a conservative 9-bp in-frame deletion. As expected, this mutation was found in the six genomes identified as lineage P.1. We found only one nonsense mutation, located in ORF7a, at position 27,673 (Q94*).
Fig. 2.
Mutations of SARS-CoV-2 genome sequences from Ceará state, Northeast Brazil. a) Frequency of SNPs per SARS-CoV-2 genome position among the 34 genome sequences (missense SNPs with prevalence >40% were labelled). b) Frequency of SNPs in Spike protein (S) among the 34 genome sequences (SNPs with prevalence >20% were labelled).
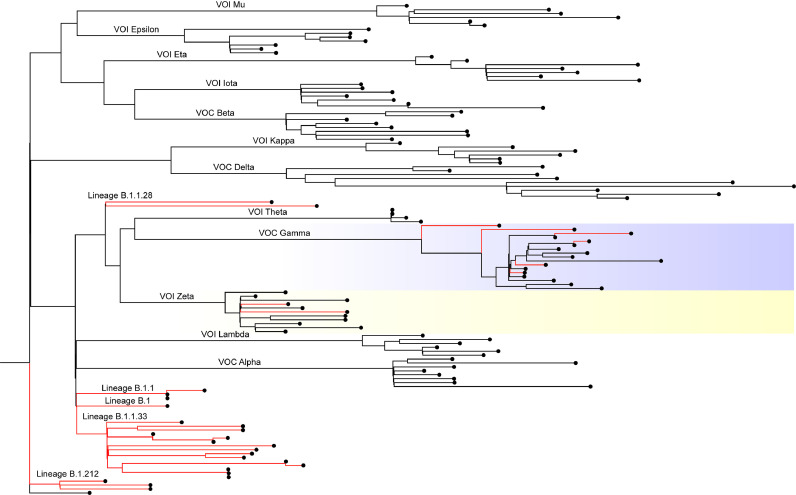
A Maximum Likelihood phylogenetic tree was constructed to confirm SARS-CoV-2 lineage classification and to show the proximity of the lineages found in the study with all variants classified as VOC or VOI that circulated from July 2020 to June 2021 (Fig. 3 ). Within the study set, only VOC Gamma and VOI Zeta were found, being classified as lineages P.1 and P.2, respectively. Apart from those sequences, lineages B.1, B.1.1, B.1.1.33, B.1.1.28 and B.1.212 were found within the dataset, which show low relationship to other VOCs and VOIs.
Fig. 3.
ML phylogenetic tree of the genomes obtained in the present study (red branches) and representatives of all VOCs and VOIs circulating during the study period. Colored highlights indicate variants that are represented within the genomes sequenced in the study.
4. Discussion
In the present study, we showed the most prevalent circulating lineages and the distribution of SARS-CoV-2 mutations in the Brazilian state of Ceará. Genome sequencing is an essential step to understand lineage dispersion and to detect mutations. The first viral genomes sequenced available in GISAID from Ceará were classified as B.1 (Candido et al., 2020), and similar results have been reported for other Brazilian states (Botelho-Souza et al., 2021; dos Santos et al., 2021). Lineage B.1 was predominant worldwide, especially in Europe, and emerged around January 2020 (Rambaut et al., 2020), contributing to the early viral epidemic dynamics in Brazil (Candido et al., 2020). Within our dataset, B.1.1.33 was the most prevalent lineage in Ceará in 2020, which was in accordance with another study, also conducted in the Northeast region of Brazil, that showed a higher prevalence of B.1.1.33 lineage (dos Santos et al., 2021). In a different study, a low prevalence of B.1.1.33 in Ceará, in the first two months of the pandemic, was reported (Resende et al., 2021). However, divergences of results are expected due to sampling size bias. The results showed occurrence of lineage P.2 in April 2020, with the highest frequency in January 2021. In Brazil, lineage P.2 was first reported in Rio de Janeiro, in October 2020 (Voloch et al., 2020), even though Lamarca et al. (2021) estimated that the origin of P.2 lineage took place in February 2020. From the results, lineage P.1 was detected in Ceará in mid-October 2020, and quickly became predominant in early 2021, being responsible for most COVID-19 infections in Brazil during the second wave (Levi et al., 2021). P.1 (VOC Gamma) was first detected in Manaus in November-December 2020, and was quickly found in other Brazilian states (Faria et al., 2021b). Recently, Lamarca et al. (2021) inferred that P.1 had its origin around August 2020, which is in accordance with our results. This suggests that lineage P.1 emerged and was not noticed earlier due to poor genomic surveillance in the country at that time. Our analysis identified genome mutations within a dataset comprising 34 samples collected from patients in Ceará, using as reference the genome of the first SARS-CoV-2 isolated in Wuhan, China. In the Spike protein, the mutation D614G was most prevalent, with 100% of frequency within the dataset. D614G was first detected in January 2020, in samples from China and Germany, but quickly became the dominant genotype throughout the world (Korber et al., 2020; Yurkovetskiy et al., 2020). Moreover, D614G has been associated with lower Ct values in infected patients, possibly indicating a higher upper respiratory tract viral load. D614G has been previously associated with ORF1ab:P4715L, with a strong allelic association, therefore, they possibly confer a fitness gain (Yang et al., 2020). Indeed, these mutations had a high prevalence among the sequenced genomes, which can indicate an increased transmissibly of the virus. Moreover, mutations P4715L and D614G were detected in South America, including Brazil, and have been correlated with higher mortality rates (Fang et al., 2021; Toyoshima et al., 2020). Among our samples, we also detected mutations that were located in the Spike protein that have been associated with a higher infectivity and evasion of immune system, such as K417T, E484K and N501Y (Harvey et al., 2021; Khan et al., 2021). E484K has also been detected in lineage P.2 (Voloch et al., 2020), and in one of the genomes classified as B.1.1.33 from July 2020, within the dataset. Some Clade 2 signature mutations, which showed a wide spread in Brazil (Candido et al., 2020), were also found in the dataset: I33T, located in ORF6, and I292T, in the N gene. Structural proteins encoded for the two regions are involved in the degradation of interferon-induced antiviral proteins (Li et al., 2020). Finally, other important mutations found with a higher prevalence among our dataset were R203K and G204R, located in the N gene. The occurrence of these SNPs was reported by Laamarti et al. (2020), who detected them in samples from all continents except Africa and Asia. Our results highlight the importance of genomic surveillance as a tool for monitoring and understanding the evolution of SARS-CoV-2. Furthermore, this work fills a gap in the knowledge about SARS-CoV-2 as it reports the early imports of lineages and the prevalence of mutations in the state of Ceará in the first two years of the COVID-19 pandemic in Brazil.
5. Conclusions
We have showed that the lineage B.1.1.33 was the most prevalent in early epidemic phase in Ceará state in 2020, and that P.1 was predominant in 2021. The mutations reported in this present study also brought up the genetic diversity of SARS-CoV-2 variants and provided evidence associated with higher transmissibility and disease severity. Our results confirm the need to sustain continuous genomic surveillance through SARS-CoV-2 sequencing in order to identify circulating lineages and to monitor the pandemic.
Funding
This study was supported by Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP – grant number 03195011/2020); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES - grant numbers 23038.001015/2020-15 and 88887.512361/2020-00).
Supplementary materials
Supplementary Table S1 – SARS-CoV-2 genome sequences from the state of Ceará, Northeast Brazil, collected until June 2021
Supplementary Table S2 – Number of SARS-CoV-2 genome sequences used in the analysis from March 2020 to June 2021
Supplementary Table S3 – Prevalence of SNPs identified in the 34 genomes sequenced
Supplementary Table S4 – SARS-CoV-2 missense mutations found in the study dataset
Supplementary Method – The dataset for the Maximum Likelihood tree
CRediT authorship contribution statement
Francisca Andréa da Silva Oliveira: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Maísa Viana de Holanda: Conceptualization, Methodology, Writing – original draft. Luína Benevides Lima: Conceptualization, Formal analysis, Writing – original draft. Mariana Brito Dantas: Conceptualization, Methodology. Igor Oliveira Duarte: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Luzia Gabrielle Zeferino de Castro: Formal analysis, Investigation. Laís Lacerda Brasil de Oliveira: Methodology, Investigation. Carlos Roberto Koscky Paier: Methodology, Investigation. Caroline de Fátima Aquino Moreira-Nunes: Conceptualization, Investigation. Nicholas Costa Barroso Lima: Conceptualization, Formal analysis, Investigation, Writing – original draft. Maria Elisabete Amaral de Moraes: Funding acquisition, Resources, Supervision. Manoel Odorico de Moraes Filho: Funding acquisition, Resources, Supervision. Vânia Maria Maciel Melo: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. Raquel Carvalho Montenegro: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198908.
Appendix. Supplementary materials
Data Availability
Data will be made available on request.
References
- Alonge M., Soyk S., Ramakrishnan S., Wang X., Goodwin S., Sedlazeck F.J., Lippman Z.B., Schatz M.C. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S., 2010. A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho-Souza L.F., Nogueira-Lima F.S., Roca T.P., Naveca F.G., de Oliveria dos Santos A., Maia A.C.S., da Silva C.C., de Melo Mendonça A.L.F., Lugtenburg C.A.B., Azzi C.F.G., Fontes J.L.F., Cavalcante S., de Cássia Pontello Rampazzo R., Santos C.H.N., Di Sabatino Guimarães A.P., Máximo F.R., Villalobos-Salcedo J.M., Vieira D.S. SARS-CoV-2 genomic surveillance in Rondônia, Brazilian Western Amazon. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-83203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., Manuli E.R., Thézé J., Almeida L., Menezes M.T., Voloch C.M., Fumagalli M.J., Coletti T.M., da Silva C.A.M., Ramundo M.S., Amorim M.R., Hoeltgebaum H.H., Mishra S., Gill M.S., Carvalho L.M., Buss L.F., Prete C.A., Ashworth J., Nakaya H.I., Peixoto P.S., Brady O.J., Nicholls S.M., Tanuri A., Rossi Á.D., Braga C.K.V., Gerber A.L., de Guimarães A.P.C., Gaburo N., Alencar C.S., Ferreira A.C.S., Lima C.X., Levi J.E., Granato C., Ferreira G.M., Francisco R.S., Granja F., Garcia M.T., Moretti M.L., Perroud M.W., Castiñeiras T.M.P.P., Lazari C.S., Hill S.C., de Souza Santos A.A., Simeoni C.L., Forato J., Sposito A.C., Schreiber A.Z., Santos M.N.N., de Sá C.Z., Souza R.P., Resende-Moreira L.C., Teixeira M.M., Hubner J., Leme P.A.F., Moreira R.G., Nogueira M.L., Ferguson N.M., Costa S.F., Proenca-Modena J.L., Vasconcelos A.T.R., Bhatt S., Lemey P., Wu C.H., Rambaut A., Loman N.J., Aguiar R.S., Pybus O.G., Sabino E.C., Faria N.R. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Patel V.M., Coon M., Nguyen T., Land S.J., Ruden D.M., Lu X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012;3:1–9. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:1–4. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus J.G., Sacchi C., Candido Sd.da S., Claro I.M., Sales F.C.S., Manuli E.R., Silva D.B.B.da, Paiva T.M.De, Pinho M.A.B., Santos K.C.de O., Hill S.C., Aguiar R.S., Romero F., Santos F.C.P.dos, Gonçalves C.R., Timenetsky M.do C., Quick J., Croda J.H.R., Oliveira W.de, Rambaut A., Pybus O.G., Loman N.J., Sabino E.C., Faria N.R. Importation and early local transmission of COVID-19 in Brazil, 2020. Rev. Inst. Med. Trop. Sao Paulo. 2020:1–5. doi: 10.1590/S1678-9946202062030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos C.A., Bezerra G.V.B., de Azevedo Marinho A.R.R.A., Alves J.C., Tanajura D.M., Martins-Filho P.R. SARS-CoV-2 genomic surveillance in Northeast Brazil: timing of emergence of the Brazilian variant of concern P1. J. Travel Med. 2021;28:1–3. doi: 10.1093/jtm/taab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Liu S., Shen J., Lu A.Z., Wang A.K.Y., Zhang Y., Li K., Liu J., Yang L., Hu C.D., Wan J. Updated SARS-CoV-2 single nucleotide variants and mortality association. J. Med. Virol. 2021;93:6525–6534. doi: 10.1002/jmv.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Claro I.M., Candido D., Franco L.A.M., Andrade P.S., Coletti T.M., Silva C.A.M., Sales F.C., Manulli E.R., Aguiar R.S., Gaburo N., Camilo C., da C., Fraiji N.A., Crispim M.A.E., Carvalho M.P.S.S., Rambaut A., Loman N., Phybus O.G., Sabino E.C. Virological.Org; 2021. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings; pp. 1–9. [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit, Ruben J.G., Franco L.A.M., Raimundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., da Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U.G., Gaburo N., Jr, Camilo C.da C., Hoeltgebaum H., Souza W.M., Rocha E.C., Souza L.M.de, Pinho M.C.de, Araujo L.J.T., Malta F.S.V, Lima A.B.de, Silva J.do P., Zauli D.A.., Ferreira A.C.S., Schnekenberg R.P., Laydon D.J., Walker Patrick, G.T., Schluter H.M., Santos A.L.P.dos, Vidal M.S., Caro V.S.Del, Filho R.M.F., Santos H.M.dos, Aguiar R.S., Proenca-Modena J.L., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Carlos P.J., Nascimento V.H., A S.M., Bowden T.A., Pond S.L.K., Wu C.-H., Ratmann O., Ferguson N.M., Christopher D., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M.do P.S.S., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. 80-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T.C., Yeo W., Team G.C.C., Maurer-Stroh S. GISAID's role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laamarti Meriem, Alouane T., Kartti S., Chemao-Elfihri M.W., Hakmi M., Essabbar A., Laamarti Mohamed, Hlali H., Bendani H., Boumajdi N., Benhrif O., Allam L., Hafidi N.El, Jaoudi R.El, Allali I., Marchoudi N., Fekkak J., Benrahma H., Nejjari C., Amzazi S., Belyamani L., Ibrahimi A. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geodistribution and a rich genetic variations of hotspots mutations. PLoS One. 2020;15:1–18. doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarca A.P., de Almeida L.G.P., Francisco R.da S., Lima L.F.A., Scortecci K.C., Perez V.P., Brustolini O.J., Sousa E.S.S., Secco D.A., Santos A.M.G., Albuquerque G.R., Mariano A.P.M., Maciel B.M., Gerber A.L., Guimarães A.P.de C., Nascimento P.R., Neto F.P.F., Gadelha S.R., Porto L.C., Campana E.H., Jeronimo S.M.B., Vasconcelos A.T.R. Genomic surveillance of SARS-CoV-2 tracks early interstate transmission of P.1 lineage and diversification within P.2 clade in Brazil. PLoS Negl. Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2 - what do they mean? JAMA - J. Am. Med. Assoc. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Levi J.E., Oliveira C.M., Croce B.Della, Telles P., Lopes A.C.W., Romano C.M., Lira D.B., Resende A.C.M.de, Lopes F.P., Ruiz A.A., Campana G. Dynamics of SARS-CoV-2 variants of concern in Brazil, early 2021. Front. Public Heal. 2021;9:1–6. doi: 10.3389/fpubh.2021.784300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A., Lanfear R., Teeling E. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Delatorre E., Gräf T., Mir D., Motta F.C., Appolinario L.R., Paixão A.C.D.da, Mendonça A.C., da F., Ogrzewalska M., Caetano B., Wallau G.L., Docena C., Santos M.C.dos, de Almeida Ferreira J., Sousa Junior E.C., Silva S.P.da, Fernandes S.B., Vianna L.A., Souza L., da C., Ferro J.F.G., Nardy V.B., Santos C.A., Riediger I., do Carmo Debur M., Croda J., Oliveira W.K., Abreu A., Bello G., Siqueira M.M. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Front. Microbiol. 2021;11:1–14. doi: 10.3389/fmicb.2020.615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., Iwatsuki-Horimoto K., Ito M., Yamayoshi S., Loeber S., Tsuda M., Wang L., Ozono S., Butlertanaka E.P., Tanaka Y.L., Shimizu R., Shimizu K., Yoshimatsu K., Kawabata R., Sakaguchi T., Tokunaga K., Yoshida I., Asakura H., Nagashima M., Kazuma Y., Nomura R., Horisawa Y., Yoshimura K., Takaori-Kondo A., Imai M., Chiba M., Furihata H., Hasebe H., Kitazato K., Kubo H., Misawa N., Morizako N., Noda K., Oide A., Suganami M., Takahashi M., Tsushima K., Yokoyama M., Yuan Y., Tanaka S., Nakagawa S., Ikeda T., Fukuhara T., Kawaoka Y., Sato K. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021;602 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souvorov A., Agarwala R., Lipman D.J. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19:1–13. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G., O'Connor B. 1st Ed. O'Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- Voloch C.M., Jr R.S.F., Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., de Guimarães A.P.C., Mariani D., da Costa R.M. medRxiv; 2020. Genomic Characterization of a Novel SARS-CoV-2 Lineage from Rio de Janeiro, Brazil. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- WHO, 2022. Coronavirus (COVID-19) Dashboard. http://covid19.who.int/.
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.C., Chen C.H., Wang J.H., Liao H.C., Yang C.T., Chen C.W., Lin Y.C., Kao C.H., Lu M.Y.J., Liao J.C. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc. Natl. Acad. Sci. U. S. A. 2020;117:30679–30686. doi: 10.1073/pnas.2007840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.