Figure 1 Key figure. Overview of putative integration site-dependent mechanisms that can affect host protein expression.
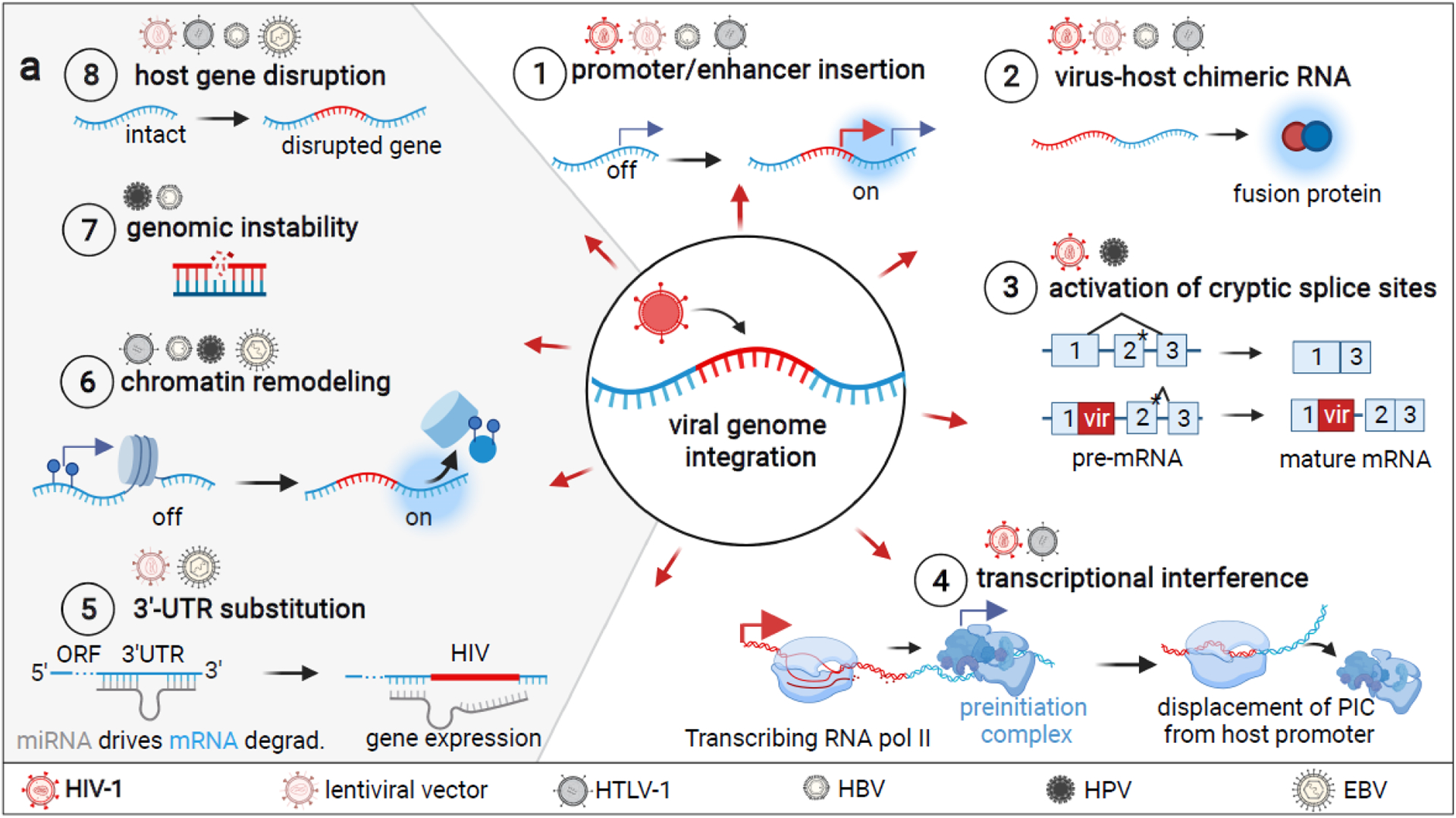
Un-demonstrated HIV-1 integration mechanisms are depicted in gray and viruses for which experimental evidence exists are depicted above each. 1) Integration of a viral promoter or enhancer can activate gene expression [6,8,9,16]. 2) Viral or host promoters may drive the transcription of chimeric virus-host RNA and translation of fusion proteins [6,16]. 3) Disruption of a splice site can result in the activation of a downstream cryptic splice site (denoted with *) that was previously disfavored for a stronger splice site [8,16]. This can result in the retention of introns in mRNA [16]. 4) Transcriptional interference of viral and host promoters can result in various scenarios in which only either cellular or viral transcription occurs [49,50], regardless of the orientation of the two promoters. Typically, it allows for transcription of the gene driven by the stronger promoter only [52]. 5) Genes can be upregulated by mRNA 3′ end substitution if a degradation-promoting 3′ mRNA end is disrupted by viral integration [42–44]. 6) Viral sequences interact with various host chromatin remodelers and may interact with other chromatin regions to impact 3D organization/accessibility [19,24–26]. 7) Integrations may entail single or double strand breaks that result in various mutations, including chromosomal translocations in the case of HBV [27,28,32]. 8) Viral integration may disrupt host genes via the insertion of premature STOP codons, poly-A sites, or frameshift mutations [23,29,32,45,64,102,103]. This figure was created with BioRender.com.
