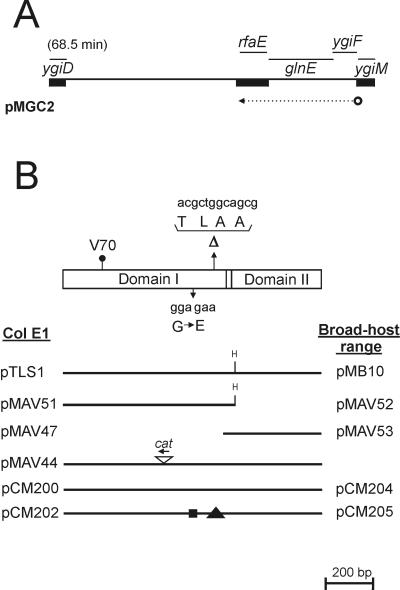
FIG. 3.
(A) Partial map of the chromosomal region cloned into pMGC2 that corresponds to the E. coli K-12 genetic map. Thick bars represent regions that were sequenced. The arrow indicates the direction of transcription of the putative ygiF-glnE-rfaE operon. (B) Map of the rfaE gene. Boxes correspond to the Domain I and Domain II regions. The positions of the various mutations are indicated: V70, the position of the Tn10 insertion in KLC2926 after the codon encoding valine 70 of the RfaE protein; ΔTLAA, the location of the 12-bp deletion in SL1102 rfaE and the corresponding amino acids; G→E, the glycine-to-glutamic acid substitution due to a base pair change (GGA to GAA). The partial maps show the various plasmids. The ColE1 vector was pEX1, and these plasmids were used in the experiments involving E. coli and S. enterica. The broad-host-range vector was pME6000, and these plasmids were used in the experiments involving R. eutrophus. The position and orientation (arrow) of the insertion of a cat cassette within the E. coli rfaE gene in pMAV44 is shown. pCM200 and pCM204 correspond to the cloned wild-type rfaE gene of S. enterica SL1027. pCM202 and pCM205 correspond to the cloned mutant rfaE543 gene of S. enterica SL1102. The box and triangle in pCM202/pCM205 denote the positions of the amino acid substitution and deletion, respectively.