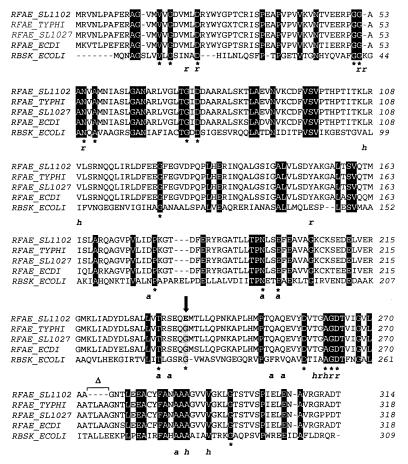
FIG. 4.
Amino acid sequencing alignment of Domain I of RfaE proteins from E. coli (RFAE_ECDI), S. enterica serovar Typhi (RFAE_TYPHI), S. enterica serovar Typhimurium strains SL11027 (RFAE_SL1027) and SL1102 (RFAE_SL1102), and E. coli ribokinase (RBSK_ECOLI). The alignment was produced with CLUSTAL W. Identical residues in all five proteins are boxed in black. ∗, residues that are also conserved in other members of the ribokinase family (45); r, a, and h, residues in the ribokinase that bind ribose (r) or ADP (a) via hydrogen bonds or that make van der Waals contacts with ribose and/or ADP (h) (for more details on the structure of ribokinase see reference 45). The arrow indicates the substitution of glycine (boxed in grey) by glutamic acid (bold) in the SL1102 RfaE mutant of SL1102. ▵, the deletion of four amino acids in the SL1102 RfaE mutant.