FIG 1.
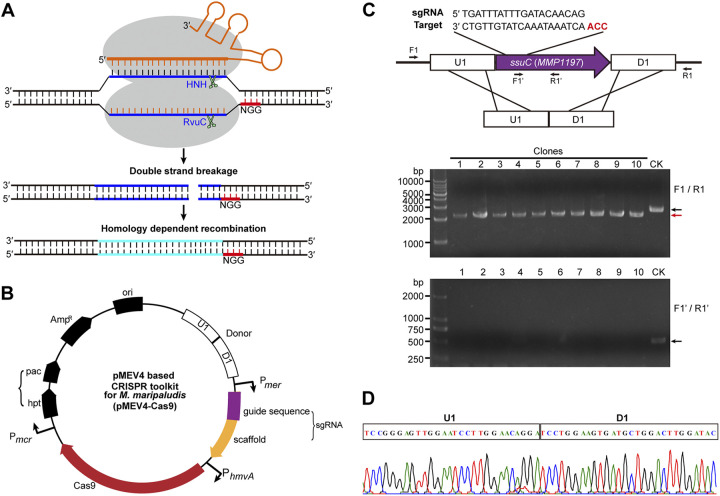
Schematic illustration of the CRISRP-Cas9 system developed in M. maripaludis and its application in the deletion of MMP1197. (A) Diagram of the Cas9-based genome editing. The Streptococcus pyogenes Cas9 and also a chimeric small guide RNA (sgRNA), which contains a 20-bp sequence derived from the host chromosome (blue) immediately upstream of a NGG PAM (red) and an 80-bp scaffold sequence (orange) to facilitate Cas9 binding, were inserted into the pMEV4 series plasmids and expressed under the respective promoters indicated in panel B. Using a donor carrying the repair templates for homologous recombination (cyan) to repair the double-strand breakages in the polyploid chromosomes, gene deletions, tagging, and multiple-/single-nucleotide mutagenesis could be generated by the Cas9-based system. HNH, histidine-asparagine-histidine endonuclease domain of Cas9. (B) Map of the pMEV4-based Cas9 editing plasmid used in M. maripaludis. Cas9 and sgRNA were expressed under the control of the indicated promoters. Pac, puromycin transacetylase for selection of the puromycin-resistant (Purr) transformants that carry the Cas9 editing system; Hpt, hypoxanthine phosphoribosyltransferase to confer a sensitivity to the base analog 8-azahypoxanthine used in curing the Cas9 editing plasmid. (C) Knockout of ssuC (MMP1197) using the Cas9 editing system. Cas9 was guided by the sgRNA that targets the ORF internal of ssuC upstream of a CGG PAM (red) to bring a DSB at the ssuC locus and a donor containing a homologous recombinant template to delete 456 bp in the ssuC ORF (purple arrow) (upper portion). PCR screening of 10 selected transformants after one-step transformation and puromycin resistance selection was performed. Primers F1/R1 for specific amplifying the genome region flanking the ssuC ORF and F1′/R1′ for amplifying the deleted region were used to detect the deleted region residuals in the polyploid chromosomes (lower portion). Black and red arrows indicate the PCR products amplified from the wild-type (lane CK) and the mutated (lanes 1 to 10) genomes, respectively. M, double-stranded DNA (dsDNA) size marker. (D) A representative sequencing result of the PCR product amplified from the genomes of the selected transformants with ssuC deletions in panel C.