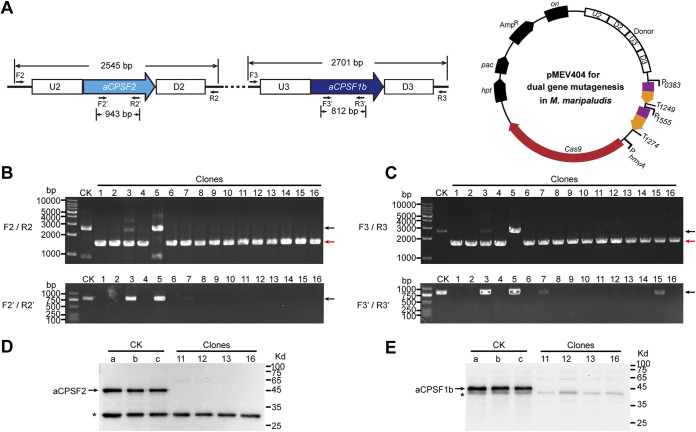
FIG 2.
Synchronous deletion of two genes in M. maripaludis using the Cas9 editing system via one-step transformation. (A) Designed schematic of synchronous deletion of the genes aCPSF2 and aCPSF1b, which encode two putative β-CASP family ribonucleases (left). Plasmid pMEV404 encodes two sgRNAs that match the ORF internal sequences of aCPSF2 and aCPSF1b (right). The sequences upstream (U2 and U3) and downstream (D2 and D3) of each target gene were concatenated on pMEV404 to construct the recombinant donor. (B and C) Sixteen puromycin-resistant transformants were randomly selected and subjected to PCRs for detecting the knockout of aCPSF2 (B) and aCPSF1b (C). The PCR primer pairs F2/R2, F2′/R2′, F3/R3, and F3′/R3′ were designed similarly to F1/R1 and F1′/R1′, as described in the legend of Fig. 1. Black and red arrows indicate the PCR products amplified from the wild-type (lane CK) and the mutated genomes (lanes 1-16), respectively. M, dsDNA size marker. (D and E) Western blot assay determined the knockout of aCPSF2 (D) and aCPSF1b (E) in the selected transformants. Arrows indicate the respective protein bands of aCPSF2 and aCPSF1b through hybridization with the antibody of each protein. Asterisks indicate a band resulting from nonspecific hybridization of the polyclonal antibody. A protein ladder with molecular weights indicated is shown at the right.