FIG 7.
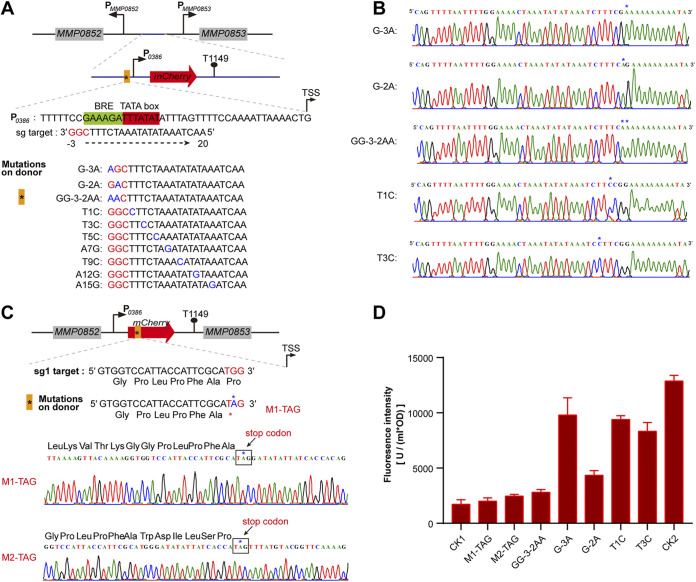
Cas9-based in situ mutagenesis of single nucleotides for determining key residues in promoter or inactivating the expression of the target gene in M. maripaludis. (A) Design schematic showing the in situ mutagenesis of single-nucleotide substitution in P0386, the promoter of MMP0386, which is fused with the reporter mCherry gene to obtain P0386-mCherry. P0386-mCherry was inserted in the intergenic region between MMP0852 and MMP08530 as in our previous study (34). An sgRNA targeting the promoter region (orange block) was expressed and the substitution of the single nucleotide (blue letters) to mutate the key residues of the promoter was introduced into the donor sequence on the pMEV4 plasmids. PAM nucleotides are shown in red. (B) Five randomly selected transformants were screened by PCRs, and the nucleotide mutations were further validated by DNA sequencing. (C) Design schematic of introducing a stop codon into the mCherry gene via Cas9-based in situ single-nucleotide substitution. An sgRNA targeting the ORF region (orange block) was expressed and the single-nucleotide substitutions (indicated by the asterisk) to introduce stop codons were generated in the donor sequence on the pMEV4 plasmids. PAM nucleotides are shown in red. Five randomly selected transformants were screened using PCRs, and the nucleotide mutations were further validated by DNA sequencing. (D) Fluorescence intensities of mCherry were assayed to determine the effects on the expression of mCherry of the single-nucleotide mutagenesis in the promoter and the introduction of a stop codon. CK1, CK2, and other symbols on the x axis indicate the parental strain S0001, S0001 carrying P0386-mCherry, and the mutation strains obtained as shown in panels A and C. Fluorescence intensities of mCherry were assayed in triplicate cultures, and averages and standard deviations are shown.