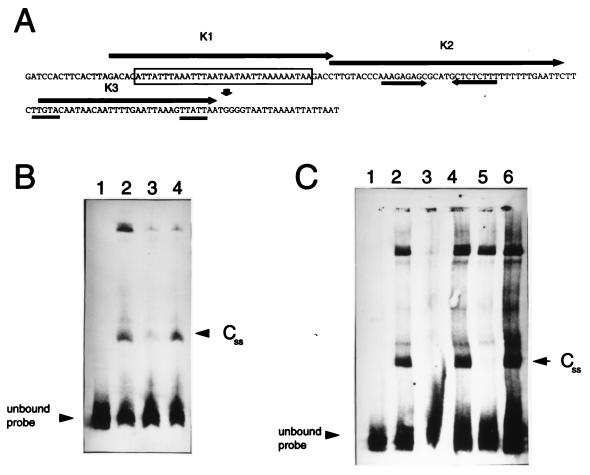
FIG. 2.
(A) Nucleotide sequence of the 157-bp p35 promoter probe used in the EMSA reactions (4). The transcription start site is indicated by the downward pointing arrow. An inverted repeat is indicated by the converging arrows. The putative promoter region is doubly underlined. A 32-nt AT tract is boxed. The arrows designated K1 through K3 encompass each of the oligonucleotides used in the EMSA competition experiments. (B) Competition of DNA binding with the flaB promoter region as competitor. Twenty-three femtomoles of the 157-bp DIG-labeled BamHI-PacI p35 promoter probe was electrophoresed with no cell-free extract added (lane 1) or with 7 μg of extract from spirochetes harvested in stationary phase (lanes 2 to 4), plus a 50 M excess of the unlabeled 157-bp BamHI-PacI p35 probe (lane 3) or a 50 M excess of the unlabeled 175-bp flaB promoter probe (lane 4). (C) Competition of DNA binding with small synthetic double-stranded oligonucleotides. Twenty-three femtomoles of the 157-bp DIG-labeled BamHI-PacI p35 promoter probe was electrophoresed with no cell-free extract added (lane 1) or with 7 μg of extract from spirochetes harvested in stationary phase (lanes 2 to 6), plus a 100 M excess of the unlabeled 157-bp BamHI-PacI probe (lane 3), a 100 M excess of K3 (lane 4), a 100 M excess of K2 (lane 5), or a 100 M excess of K1 (lane 6).