LETTER
Carbapenem resistance, mainly mediated by the production of carbapenemases, poses a serious threat to global public health (1). KPC-2 and IMP-4 serve as two important representatives of carbapenemases that have been commonly found on transmissible plasmids in various bacterial species (2–4). There were limited reports of the co-production of KPC-2 and IMP-4, only in clinical Klebsiella pneumoniae and Klebsiella oxytoca isolates in China (5–8). Here, we characterize a multidrug-resistant (MDR) Aeromonas caviae isolate in China, and report for the first time the simultaneous presence of blaKPC-2 and blaIMP-4, carried by two new types of plasmids in this species.
A. caviae strain SCLZS52 was isolated from the influx mainstream of the wastewater treatment plant of the affiliated hospital of Southwest Medical University, in August 2019, in Sichuan, China. Antimicrobial susceptibility testing was performed using the broth microdilution method and was interpreted according to Clinical and Laboratory Standards Institute documents M45 (9). SCLZS52 was resistant to meropenem, cefotaxime, cefoxitin, ciprofloxacin, and gentamicin, intermediate to tetracycline, and susceptible to tigecycline, amikacin, and chloramphenicol. It was subjected to whole genome sequencing (WGS) by using both the MinION and Illumina HiSeq 2000 sequencers. The assembly and bioinformatic analyses of the genome were performed as previously described (10). WGS data revealed that the SCLZS52 belongs to A. caviae, and it is comprised of a 4,718,963-bp circular chromosome and eight plasmids ranging from 4,076 bp to 113,450 bp in size (Table S1 in the supplemental material). SCLZS52 has 27 known acquired antimicrobial resistance genes (ARGs) mediating multidrug resistance, including two carbapenemase-encoding genes blaKPC-2 and blaIMP-4 located on two different plasmids (Table S1). Conjugation experiments were carried out using Escherichia coli strains J53 and EC600 as recipients (10, 11). However, no transconjugant was obtained after repeated attempts, suggesting that both carbapenemase determinants were not transferable, which was consistent with that no conjugative elements were detected on the plasmids.
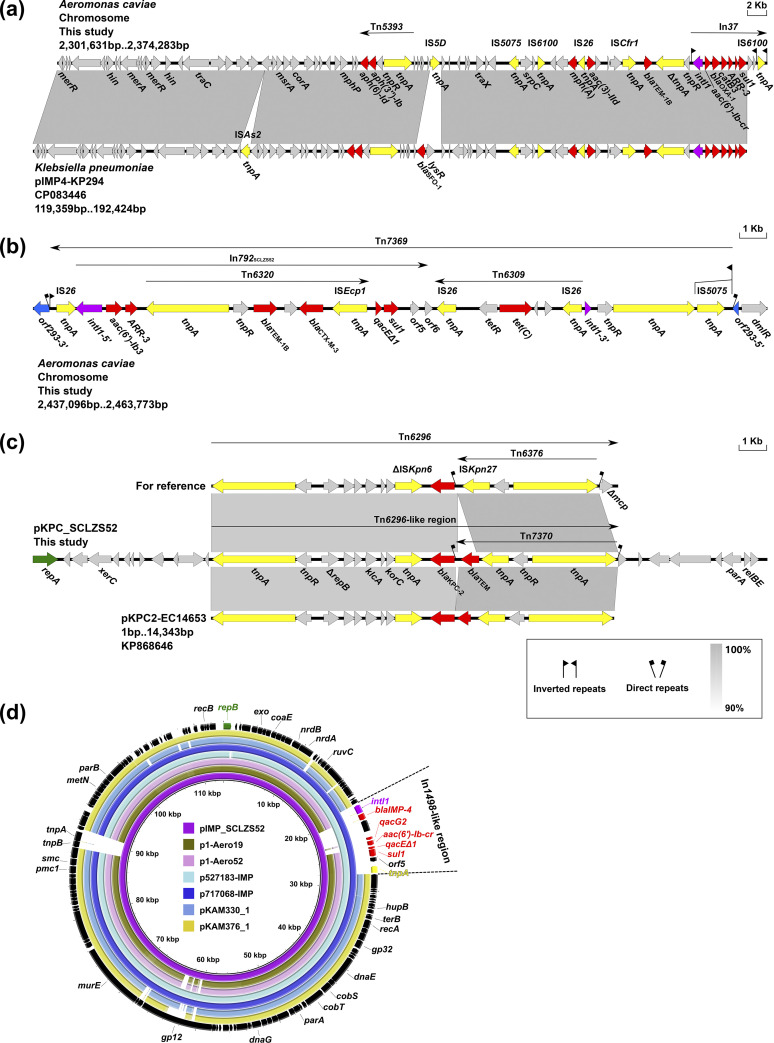
Twenty ARGs were located on the chromosome of SCLZS52, which are mainly clustered in two MDR regions, designated MDR-1 and MDR-2 (Fig. S1 in the supplemental material; Fig. 1a and b). The 41-kb MDR-1 shows >99.9% identity at 98% coverage to that of plasmids from Klebsiella, such as pIMP4-KP294 (CP083446, patient, China, 2020) and pKP1814-1 (KX839207, patient, China, 2011), suggesting that SCLZS52 may capture this segment from Klebsiella plasmids, most likely by homologous recombination (Fig. 1a). The 24-kb MDR-2 is sequentially organized as an In792 with an IS26-mediated interruption at intI1 and an insertion of Tn6320 downstream of its gene cassette, an intact Tn6309, and a core transposition module tnpAR-res with an IS5075-disrupted IRL (inverted repeat left) (Fig. 1b). The complex chimera structure is further identified as a novel transposon designated Tn7369 by the Transposon Registry. Tn7369 splits orf293 into two separate parts, leaving 6-bp direct repeats (DRs; target site duplication signals for transposition, TTCATA). BLASTn analysis revealed that Tn7369 is not common outside of SCLZS52 and its prevalence remains unclear.
FIG 1.
Genetic features of SCLZS52. (a) Comparison of the chromosomal MDR-1 of SCLZS52 with the corresponding region on plasmid pIMP4-KP294. Genes are denoted by arrows. Resistance genes, mobile genetic elements and intI1 are indicated in red, yellow, and purple, respectively. Regions of >90% homology are indicated by gray shadings. Δ represents truncated genes or mobile genetic elements. (b) The configuration of chromosomal MDR-2 region of SCLZS52. The interrupted gene orf293 is marked in blue. (c) Organization of the plasmid pKPC_SCLZS52 and comparison to related regions. The repA is indicated in green. (d) Comparison of pIMP_SCLZS52 with similar plasmids. pIMP_SCLZS52 was used as the reference. Dark arrows at the outer circles show open reading frames from pIMP_SCLZS52, with repB marked in green. Resistance genes, mobile genetic elements and intI1 in the accessory module are highlighted by colored arrows. Gaps indicate regions that were missing in the respective plasmid compared to pIMP_SCLZS52. Plasmids from inside to outside are pIMP_SCLZS52 (CP091177), p1_Aero19 (CP068233), p1_Aero52 (CP066814), p527183-IMP (MN961666), p717068-IMP (MN629346), pKAM330_1 (AP023399), and pKAM376_1(AP024403).
The plasmid pKPC_SCLZS52 (CP091179) is 26,128 bp in size, carries blaTEM in addition to blaKPC-2, and could not be assigned into any known incompatibility group. The deduced replication protein RepA belongs to the PriCT_1 superfamily (PF08708), and matches RepA proteins of two Aeromonas plasmids (WP_171281265 and WP_139750798) with >96.66% amino acid identity at >99% coverage. The backbone of pKPC_SCLZS52 had only 30% coverage (76.13% nucleotide identity) to its closest match plasmid unnamed2 (CP083946) from Aeromonas hydrophila, indicating that pKPC_SCLZS52 is a novel type of plasmid carrying blaKPC-2. In pKPC_SCLZS52, blaKPC-2 is contained in a Tn6296-like structure, wherein a novel transposon designated Tn7370, instead of Tn6376 in Tn6296, is located upstream of blaKPC-2, and the terminal Δmcp is deleted (Fig. 1c). Tn7370 is a Tn3-derived transposon with an insertion of ISKpn27 upstream of blaTEM. By BLASTn, the closest match of the blaKPC-2 region of pKPC_SCLZS52 is that of plasmid pKPC2-EC14653 (98% coverage, 98.27% identity) from Enterobacter cloacae (KP868646, patient, China, 2014), except for a 127-bp deletion between blaKPC-2 and blaTEM in the latter case, indicating a common origin of them.
The plasmid pIMP_SCLZS52 (CP091177) is 113,450 bp, wherein blaIMP-4 is contained in an In1498-like class I integron, which differed from In1498 mainly by insertion of a ltrA (encoding a putative retron-type RNA-directed DNA polymerase) downstream of blaIMP-4 and an IS6100 of the 3′-CS (3′ conserved segment). pIMP_SCLZS52 encodes a replication protein RepB of the Rep_3 superfamily (pfam10134) that does not belong to any known incompatibility group. Outside of the replication module, a cluster of genes encoding putative phage proteins are scattered in the remaining 112.1-kb region of the pIMP_SCLZS52 (Table S2 in the supplemental material). Of them, 39 genes are homologous to those of the Pseudomonas phage nickie (MG018927, wastewater, Denmark). The complete sequences of pIMP_SCLZS52 match six Aeromonas plasmids from humans and the environment with >96.53% nucleotide identity at 86–97% coverage (Fig. 1d), which constitutes a novel group of plasmids comprising a relatively conserved backbone and an accessory module carrying different ARGs, including blaIMP-4 (Fig. 1d, Fig. S2).
In conclusion, this study characterized the genomic features of an MDR A. caviae isolate, which harbors a novel type of plasmid carrying blaKPC-2 and a phage-like plasmid carrying blaIMP-4. Our work may shed new insights into the high plasticity of mobile genetic elements as vehicles in mediating the dissemination of ARGs.
Data availability.
Complete sequences of the chromosome and plasmids of SCLZS52 were deposited in GenBank under accession numbers CP091176-CP091184.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (31900125), Scientific and technological project in Sichuan Province (22MZGC0069), the Joint Funds of the Luzhou and Southwest Medical University Natural Science Foundation (2019LZXNYDJ47). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaoyi Dai, Email: daixiaoyi@swmu.edu.cn.
Luhua Zhang, Email: zhluhua@swmu.edu.cn.
Mariagrazia Perilli, University of L'Aquila.
REFERENCES
- 1.van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, Liu L, McNally A, Zong Z. 2019. Coexistence of three blaKPC-2 genes on an IncF/IncR plasmid in ST11 Klebsiella pneumoniae. J Glob Antimicrob Resist 17:90–93. doi: 10.1016/j.jgar.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Zhang W, Schwarz S, Wang C, Liu W, Chen F, Luan T, Liu S. 2019. Characterization of a blaIMP-4-carrying plasmid from Enterobacter cloacae of swine origin. J Antimicrob Chemother 74:1799–1806. doi: 10.1093/jac/dkz107. [DOI] [PubMed] [Google Scholar]
- 4.Dolejska M, Papagiannitsis CC, Medvecky M, Davidova-Gerzova L, Valcek A. 2018. Characterization of the complete nucleotide sequences of IMP-4-encoding plasmids, belonging to diverse Inc families, recovered from Enterobacteriaceae isolates of wildlife origin. Antimicrob Agents Chemother 62:e02434. doi: 10.1128/AAC.02434-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Cao W, Zhu X, Chen Z, Li L, Zhang B, Wang B, Tian L, Wang F, Liu C, Sun Z. 2012. Characterization of a novel Klebsiella pneumoniae sequence type 476 carrying both blaKPC-2 and blaIMP-4. Eur J Clin Microbiol Infect Dis 31:1867–1872. doi: 10.1007/s10096-011-1512-7. [DOI] [PubMed] [Google Scholar]
- 6.Mendes RE, Bell JM, Turnidge JD, Yang Q, Yu Y, Sun Z, Jones RN. 2008. Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob Agents Chemother 52:798–799. doi: 10.1128/AAC.01185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Z, Yu T, Qi Y, Ji S, Shen P, Yu Y, Chen Y. 2011. Coexistence of plasmid-mediated KPC-2 and IMP-4 carbapenemases in isolates of Klebsiella pneumoniae from China. J Antimicrob Chemother 66:2670–2671. doi: 10.1093/jac/dkr330. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Yuan M, Chen H, Chen X, Jia Y, Zhu X, Bai L, Bai X, Fanning S, Lu J, Li J. 2017. First report of Klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4, and KPC-2 carbapenemases. Antimicrob Agents Chemother 61:e00877. doi: 10.1128/AAC.00877-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI. 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed. CLSI guideline M45. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Qiu Y, She J, Wang X, Dai X, Zhang L. 2021. Genomic characterization of a Proteus sp. strain of animal origin co-carrying blaNDM-1 and lnu(G). Antibiotics (Basel) 10:1411. doi: 10.3390/antibiotics10111411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang B, Chang J, Cao L, Luo Q, Xu H, Lyu W, Qian M, Ji X, Zhang Q, Xia X, Yang H. 2019. Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol 19:1–12. doi: 10.1186/s12866-019-1454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00840-22-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Data Availability Statement
Complete sequences of the chromosome and plasmids of SCLZS52 were deposited in GenBank under accession numbers CP091176-CP091184.