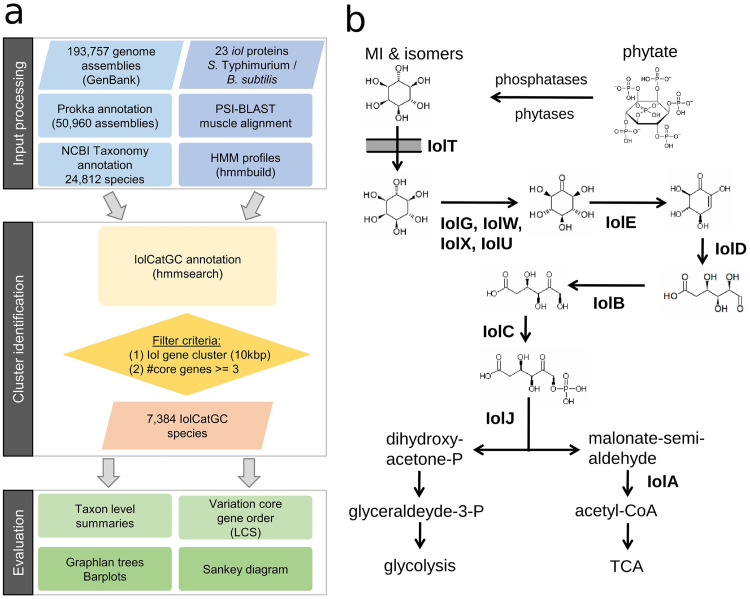
FIG 1.
Searching for IolCatGCs. (a) The workflow depicts the bioinformatic strategy applied here to perform a comprehensive prediction of all bacteria able to degrade MI. Starting with the processing of 193,757 input genome assemblies, filtering resulted in the identification of an iol gene cluster termed IolCatGC whose prevalence was then evaluated at all taxonomic levels of bacteria. (b) A schematic pathway of inositol catabolism in Gram-positive and negative bacteria is shown. Enzymes catalyzing the degradation of MI to glyceraldehyde and acetyl-CoA are indicated. IolT, transporter; IolG, IolW, IolX, IolU, IolJ, dehydrogenases of inositol and its stereoisomers; IolE, dehydratase; IolD, 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione hydrolase; IolB, isomerase; IolC, biphosphate aldolase; IolA, malonate-semialdehyde dehydrogenase; TCA, tricarboxylic acid cycle.