FIG 1.
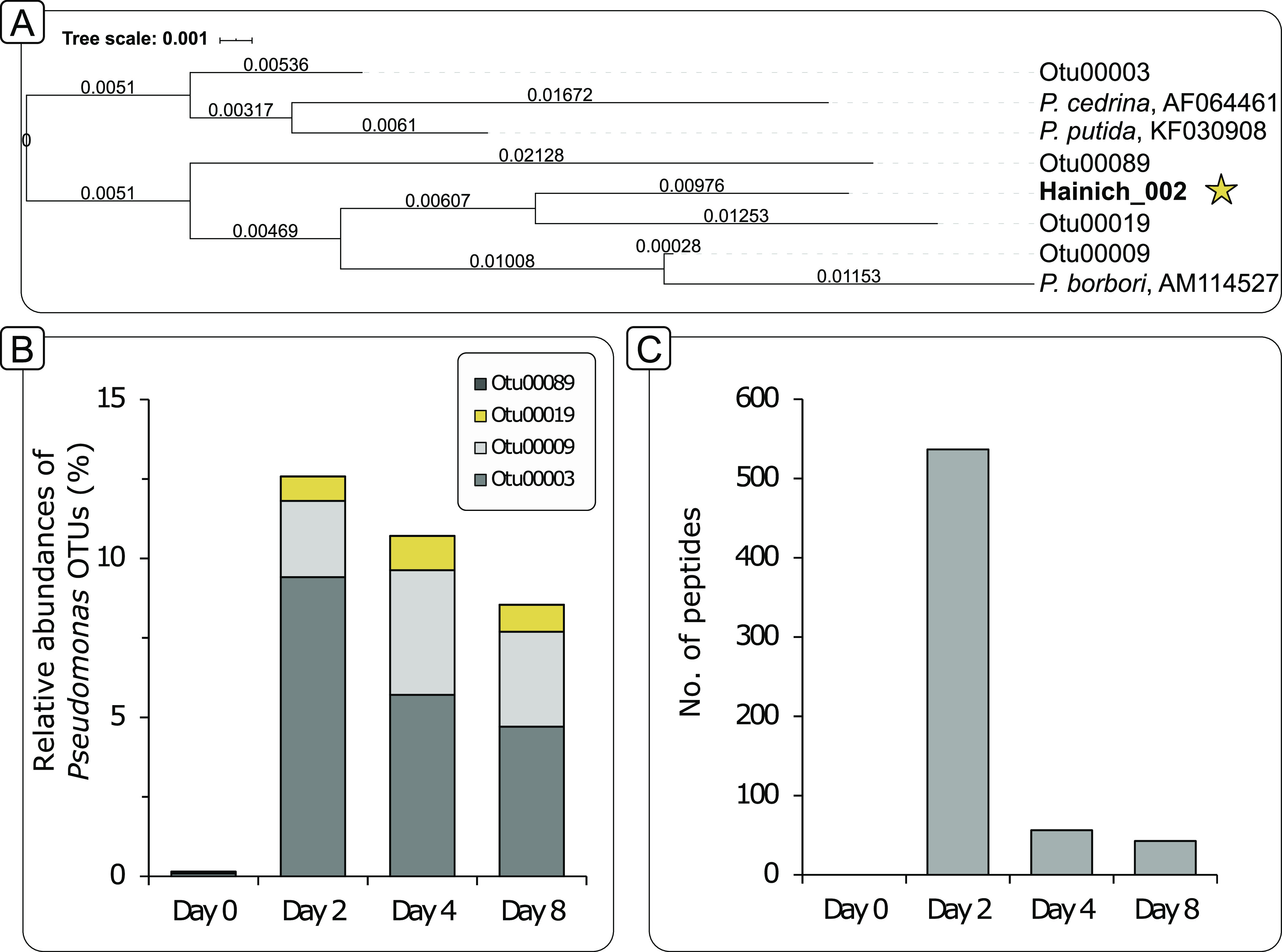
Abundances of Pseudomonas on DNA and peptide level. (A) Phylogenetic tree based on 16S rRNA gene sequences of the most abundant Pseudomonas OTUs as well the isolate Hainich_002 that was used for necromass generation (star symbol). The tree was calculated using the arb neighbor-joining method (1000 bootstraps) within arb (81, 82). Closely related Pseudomonas species were added as references. (B) Relative abundances of Pseudomonas-related OTUs increase after necromass was added on day 0 and then progressively decrease throughout the incubation. Likely, also members of the genus Pseudomonas partake in the initial degradation of Pseudomonas-derived necromass. OTU00019, the most closely related OTU to the isolate Hainich_002 is highlighted in yellow. (C) Number of peptides associated with the genus Pseudomonas within the original groundwater at day 0, as well as the 12C labeled incubations over time. Unlike at the DNA level, most Pseudomonas-related peptides presumably stem from the added necromass.
