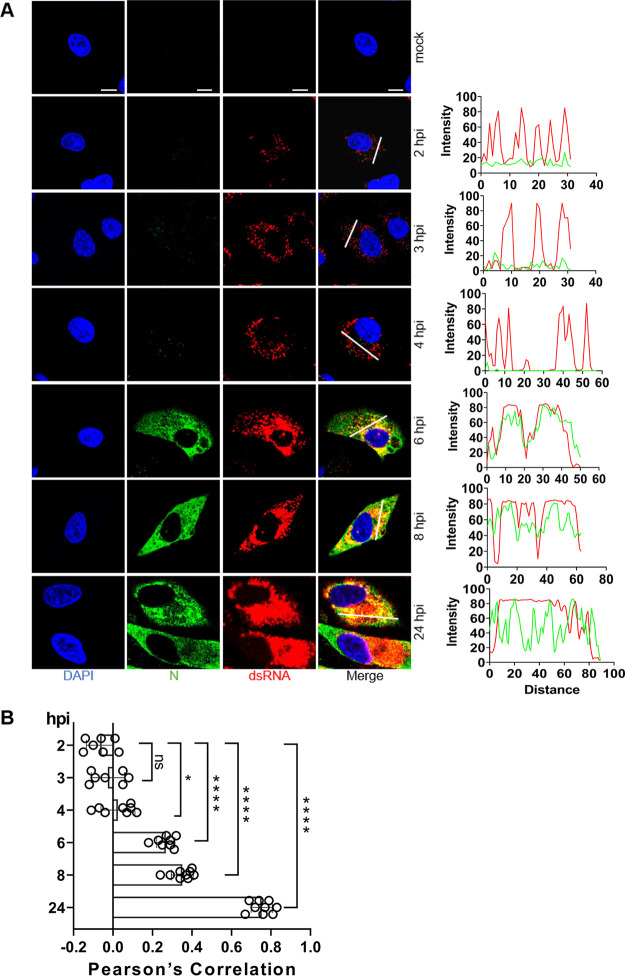
FIG 5.
Detection of dsRNA in SARS-CoV-2-infected cells. (A) A549-ACE2 cells were mock infected or infected with SARS-CoV-2 (MOI = 2), fixed at 2-, 3-, 4-, 6-, 8-, and 24-h postinfection (hpi), and costained with rabbit anti-SARS nucleocapsid (N) protein pAb and mouse anti-dsRNA monoclonal antibody (MAb). Staining was carried out with Alexa Fluor 488-conjugated goat anti-rabbit Ab (green) and Alexa Fluor 555-conjugated goat anti-mouse Ab (red). Cell nuclei were stained with DAPI (blue) and examined by confocal microscopy; scale bars represent 10 μm. The intensity distribution describes the timing of expression of the viral RNA and N proteins for specific fluorescence along the indicated line. (B) Pearson’s correlation was used to analyze changes to dsRNA and N over time. One-way analysis of variance (ANOVA) was used for multiple comparisons between different times among the viral proteins in GraphPad Prism 8.4.3 software. ****, P < 0.0001; *, P < 0.1; ns, no significance.