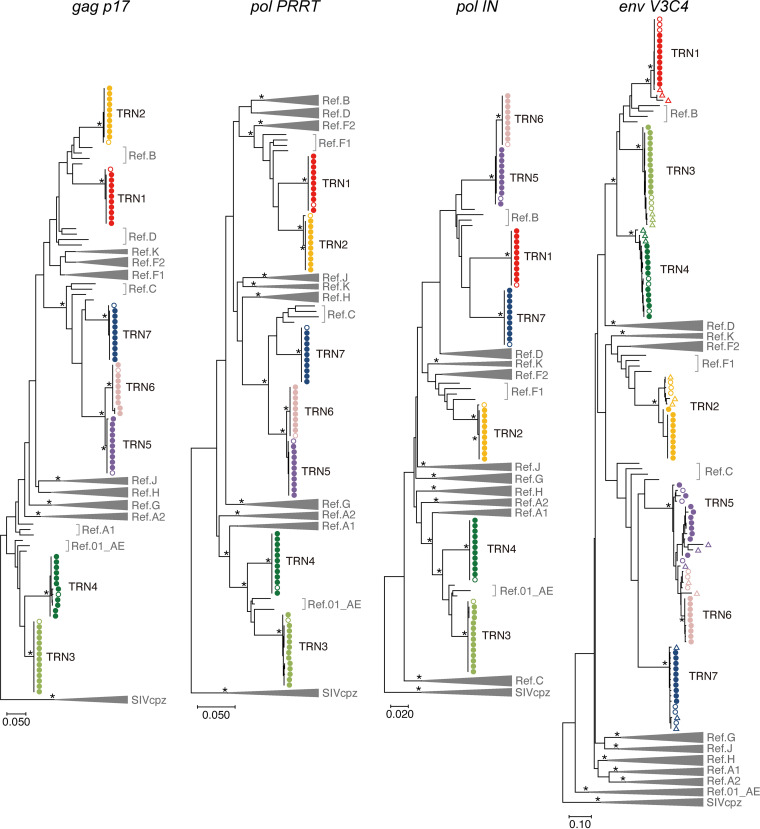
FIG 1.
Phylogenies of the RF sequences identified by nanopore sequencing and Sanger sequencing. Maximum likelihood trees with the GTR model are shown for four regions: gag p17 (positions 790 to 1185 of HXB2), pol PRRT (2253 to 3269), pol IN (4230 to 5093), and env V3C4 (7114 to 7589). The reference sequences (Ref.) of HIV-1 major subtypes (A1, A2, B, C, D, F1, F2, G, H, J, K, and CRF01_AE) and three SIVcpz strains are indicated in the tree. The three SIVcpz sequences are used as outliers (GenBank no. DQ373064, DQ373063, and EF535994 for SIVcpzLB7, SIVcpzMB66, and SIVcpzMB897, respectively). Bootstrap values were calculated by 500 replicates. Branches with bootstrap values of at least 0.95 are highlighted with asterisks. The viral full-genome sequences (closed circles) identified by nanopore sequencing and the viral RNA (open circles) and proviral DNA sequences (triangles) identified by Sanger sequencing are indicated.