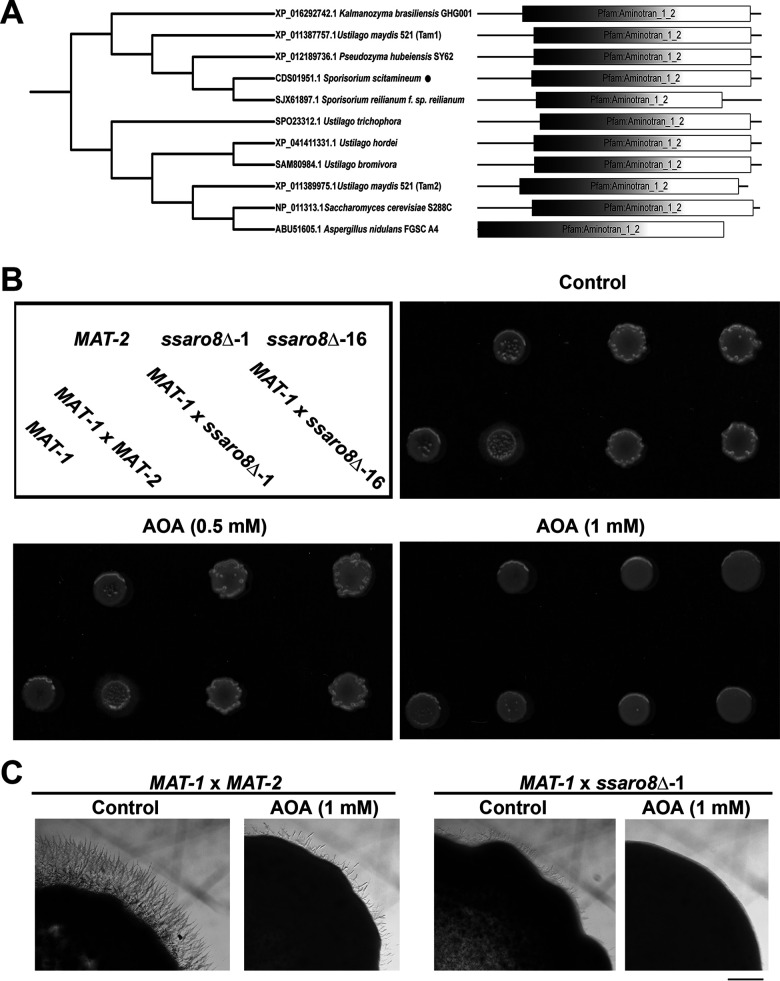
FIG 2.
Identification and characterization of SsAro8. (A) Phylogenetic analysis of orthologous Aro8 proteins in yeast and fungi. Amino acid sequences were aligned using ClustalW 2.0 and analyzed by maximum likelihood methods with 1,000 bootstrap replications. Accession number for each protein used in this analysis was presented, followed by the corresponding species/strain name. S. scitamineum SsAro8 was denoted by a solid dot. Domain prediction was performed using SMART (http://smart.embl.de/). (B) Mating assay with the sporidia of wild-type MAT-1, respectively, with the wild-type MAT-2, ssaro8Δ-1, and -16 sporidia. The mixed sporidia of opposite mating type were inoculated on PDA, with or without AOA, and allowed to grow for 3 days before photographing. (C) Observation of fungal filamentation under stereomicroscopy, at 3 dpi. Bar = 1 mm.