Figure 4.
Quantitative effects of Tet genes on transcriptomes of advanced cell types
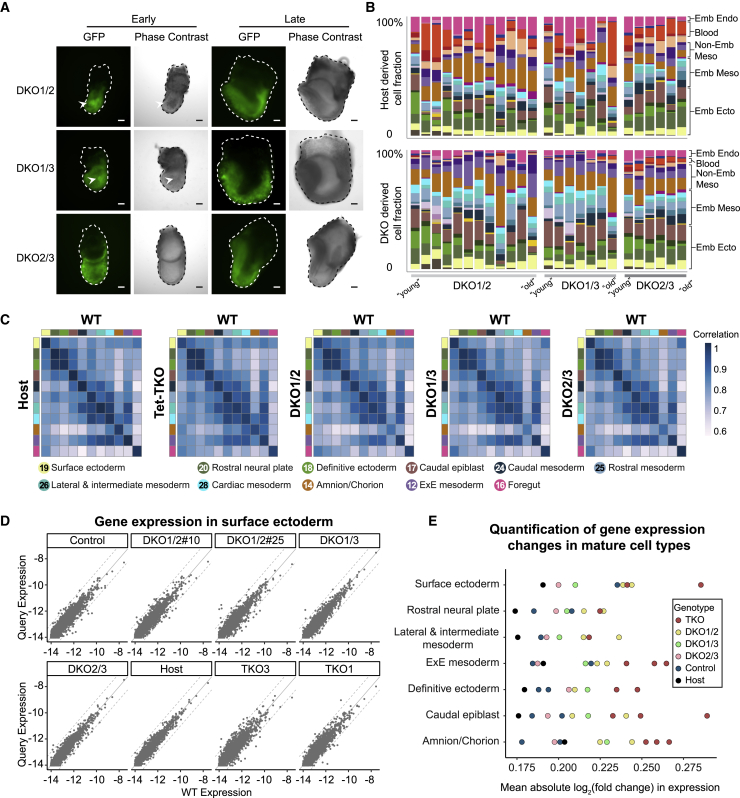
(A) Representative images of E7.5 Tet-DKO mixed chimera embryos generated by injection of GFP-labeled mESCs into 2N blastocysts. Arrowheads showing cells protruding into the cavity in early-stage DKO1/2 and DKO1/3 chimeric embryos. Scale bars, 100 μm.
(B) Cell-type composition per embryo as calculated in Figure 2E. Fraction of cell types contributed by host and by Tet-DKO-derived cells for the same embryo are shown.
(C) Transcriptional similarity (correlation of gene expression) of advanced cell types between host, Tet-TKO, DKO1/2, DKO1/3, and DKO2/3 cells from mixed chimeras and WT atlas.
(D) Scatter plot showing gene expression of surface ectoderm in lines of control and mutants compared with WT. Dashed lines indicate a 2-fold change.
(E) A summary chart showing average differential gene expression (log2 of fold change) in different cell types, each compared with the corresponding WT profile (STAR Methods). Number of embryos: 13 DKO1/2, 8 DKO1/3, 6 DKO2/3, 37 Tet-TKO, 16 Ctrl, and 45 Host embryos.
See also Figure S5.