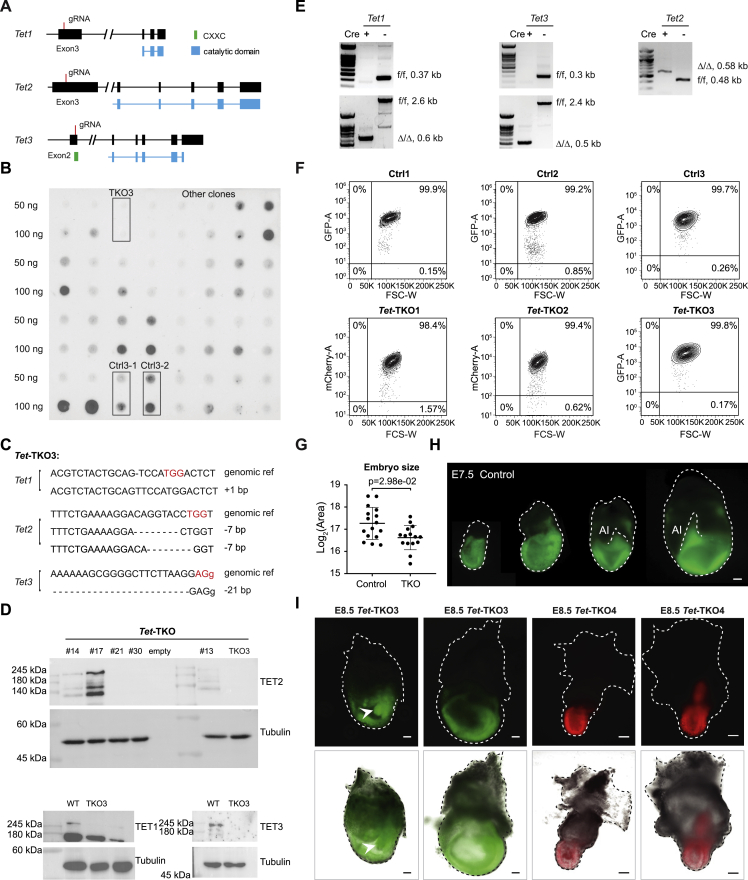
Figure S1.
Generation of fluorescently labeled Tet-TKO mESCs clones and embryos, related to Figure 1
(A) Targeting strategy using CRISPR/Cas9-mediated genome editing and 3 gRNAs against Tet genes for generation of Tet KO mESC clone. Exons and conserved domains were shown in black and green/blue boxes, introns marked in lines.
(B) Screening for clones with reduced 5hmC intensity after gene targeting using dot blot. Tet-TKO3 and Ctrl3 mESCs lines were marked in rectangles.
(C) DNA sequencing validation for Tet-TKO3 mESCs clone. Protospacer adjacent motif (PAM) sequences are marked in red. Intron sequences are shown in lower-case letters.
(D) Western blot validation for Tet-TKO3 mESCs clone. Following confirmation for lack of expression of TET2 protein, TKO3 clone was further validated for the lack of TET1 and TET3 protein expression.
(E) Generation of Tet-TKO4 mESCs clone. Tet triple floxed mESCs were derived from blastocysts followed by Cre recombinase treatment (STAR Methods). Validation of the deleted alleles was performed by PCR. f/f, homozygous floxed allele, Δ/Δ, homozygous deleted allele. Sizes of each allele were indicated on the right.
(F) Flow cytometric analysis of reporter activity for day 5 (Ctrl3 and Tet-TKO3) and day 8 (Ctrl1, Ctrl2, Tet-TKO1, and Tet-TKO2) embryoid bodies.
(G) Embryo size (extraembryonic mesodermal part excluded, log2 of μm2) comparisons between E7.5 control and Tet-TKO whole-embryo chimeras. Wilcoxon-Mann-Whitney test, two-tailed. Data are represented as mean ± SD.
(H) Representative images of E7.5 control whole-embryo chimeras. Scale bars, 100 μm.
(I) Representative images of E8.5 Tet-TKO3 and Tet-TKO4 whole-embryo chimeras. Scale bars, 100 μm.