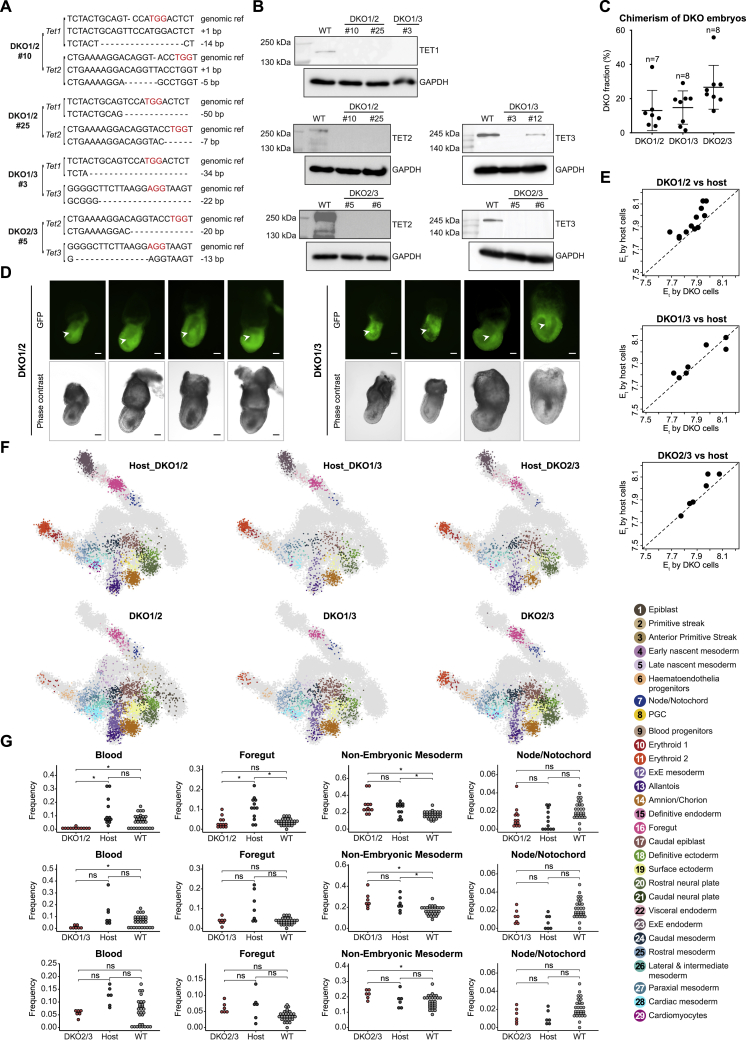
Figure S5.
Morphological and molecular perturbations in Tet-DKO mixed chimera embryos, related to Figure 4
(A) DNA sequencing validation for DKO clones after genome editing. PAM sequences are marked in red.
(B) Western blot validation of Tet-DKO mESCs clones. DKO1/2#10, DKO1/2#25, DKO1/3#3, and DKO2/3#5 were selected for subsequent chimera assays.
(C) Flow cytometric analysis for degree of chimerism for each embryo. Number of embryos shown for each genotype. Data are represented as mean ± SD.
(D) Representative images of E7.5 DKO1/2 and DKO1/3 mixed chimeric embryos. Arrowheads show the accumulation of cells inside the amniotic cavity. Embryos orientated with posterior to the right (anterior side faces up for the last DKO1/3 embryo). Scale bars, 100 μm.
(E) Comparison of Et calculated for each DKO mixed chimera embryo using both DKO- and host-derived cells.
(F) 2D-projection of transcriptome profiles onto the WT atlas (gray background) of knockout-derived cells and host-derived cells obtained from DKO1/2, DKO1/3, and DKO2/3 mixed chimera embryos, respectively. Single cells are colored by projected atlas cell type.
(G) Contribution of DKO1/2, DKO1/3, and DKO2/3 mutant cells to blood, foregut, non-embryonic mesoderm, and node/notochord as calculated in Figure 2F. Medians of frequencies were compared after downsampling of each embryo to 90 cells. Wilcoxon-Mann-Whitney rank sum test, two-tailed. ns, not significant; ∗, q value < 0.05 (Benjamini-Hochberg procedure).