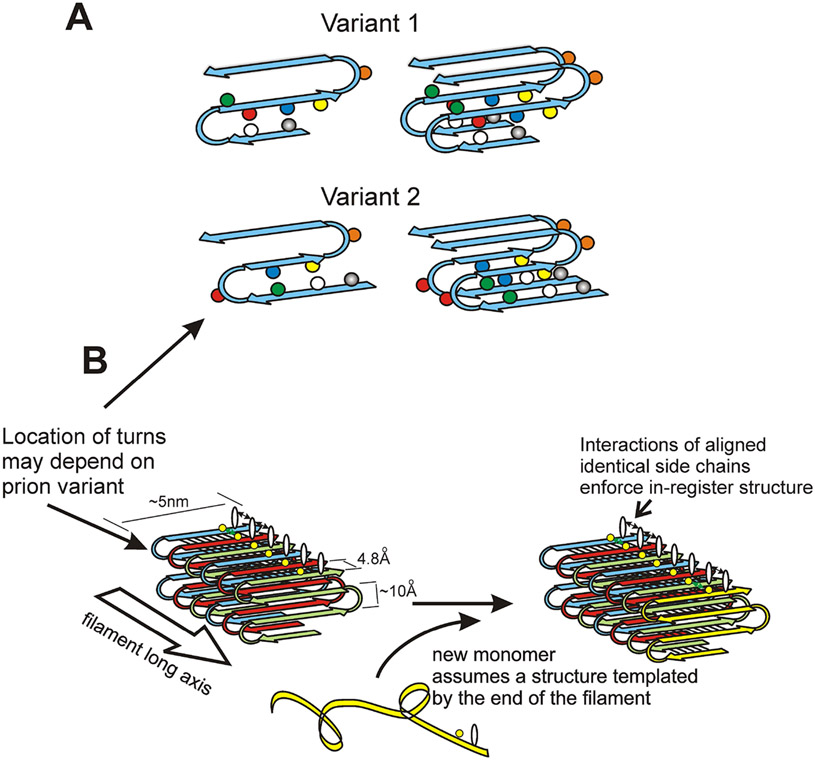
Fig. 5. Structure of the prion amyloids suggests a mechanism of conformation templating.
A. It is proposed that different prion variants have the folds of the β-sheet (turns in the molecules) in different locations (Wickner et al. 2008; Wickner et al. 2013). B. Molecules with the folded in-register parallel β-sheet architecture have rows of identical side chains along the long axis of the filament. Positive interactions (hydrogen bonds or hydrophobic interactions) between the identical side chains maintain the in-register architecture. In order to have those same positive interactions, the new unstructured prion domain joining the end of the filament must adopt a structure in which its turns are in the same location as those in the molecules already in the filament. This explains how prion proteins can template their conformation (Wickner et al. 2008). Modified from Shewmaker et al., 2006.