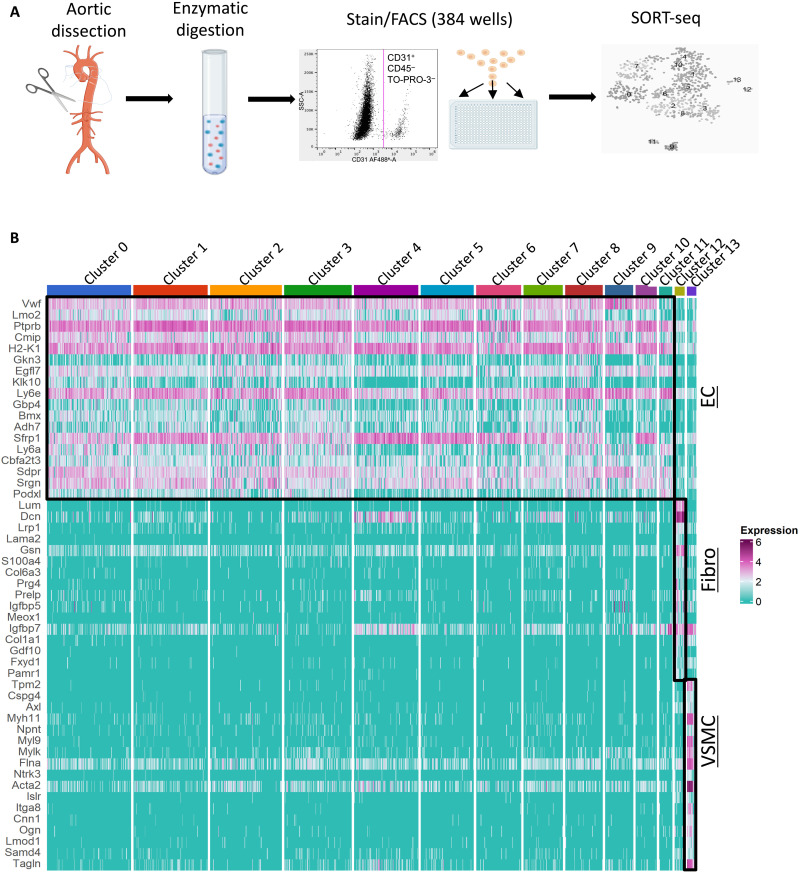
Fig. 4. scRNA-seq identification of EC subsets.
(A) Schematic overview of scRNA-seq workflow. Jag1ECKO and control (Jag1ECWT) mouse aortas were dissected from the aortic root to the iliac artery and enzymatically digested to generate single-cell suspensions. After antibody staining, CD31+, CD45−, and TO-PRO-3− cells were sorted into 384-well plates using FACS. scRNA-seq libraries were generated and sequenced using the SORT-seq method. (B) Aortas from Jag1ECKO and control mice were analyzed by FACS of CD31+ CD45− cells coupled to scRNA-seq. Heatmap showing the most highly differentially expressed genes for each cluster. Twelve distinct clusters (0 to 11) defined as ECs exhibit common expression of several markers including the canonical markers Vwf and Cdh5. Cluster 13 expresses several canonical VSMC markers including Myh11, Acta2, and Cnn1 and were therefore defined as VSMCs. Cluster 12 cells present higher expression of collagens/collagen-binding proteins (Col1a1, Dcn, and Lum), as well as reduced expression of VSMC-related proteins (Myh11 and Cnn1), and were therefore defined as fibroblasts. Fibro, fibroblasts.